Voting-based cancer module identification by combining topological and data-driven properties
- PMID: 23940583
- PMCID: PMC3734239
- DOI: 10.1371/journal.pone.0070498
Voting-based cancer module identification by combining topological and data-driven properties
Erratum in
- PLoS One. 2014;9(4):e96883
Abstract
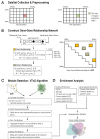
Recently, computational approaches integrating copy number aberrations (CNAs) and gene expression (GE) have been extensively studied to identify cancer-related genes and pathways. In this work, we integrate these two data sets with protein-protein interaction (PPI) information to find cancer-related functional modules. To integrate CNA and GE data, we first built a gene-gene relationship network from a set of seed genes by enumerating all types of pairwise correlations, e.g. GE-GE, CNA-GE, and CNA-CNA, over multiple patients. Next, we propose a voting-based cancer module identification algorithm by combining topological and data-driven properties (VToD algorithm) by using the gene-gene relationship network as a source of data-driven information, and the PPI data as topological information. We applied the VToD algorithm to 266 glioblastoma multiforme (GBM) and 96 ovarian carcinoma (OVC) samples that have both expression and copy number measurements, and identified 22 GBM modules and 23 OVC modules. Among 22 GBM modules, 15, 12, and 20 modules were significantly enriched with cancer-related KEGG, BioCarta pathways, and GO terms, respectively. Among 23 OVC modules, 19, 18, and 23 modules were significantly enriched with cancer-related KEGG, BioCarta pathways, and GO terms, respectively. Similarly, we also observed that 9 and 2 GBM modules and 15 and 18 OVC modules were enriched with cancer gene census (CGC) and specific cancer driver genes, respectively. Our proposed module-detection algorithm significantly outperformed other existing methods in terms of both functional and cancer gene set enrichments. Most of the cancer-related pathways from both cancer data sets found in our algorithm contained more than two types of gene-gene relationships, showing strong positive correlations between the number of different types of relationship and CGC enrichment [Formula: see text]-values (0.64 for GBM and 0.49 for OVC). This study suggests that identified modules containing both expression changes and CNAs can explain cancer-related activities with greater insights.
Conflict of interest statement
Figures


 -values of pathways and red bars indicate the overlap
-values of pathways and red bars indicate the overlap  -values between the pathway and GBM driver genes. Black vertical bars show
-values between the pathway and GBM driver genes. Black vertical bars show  -value threshold, 0.05, and the width of the horizontal bars depends on
-value threshold, 0.05, and the width of the horizontal bars depends on  (
( -value). (C) Red bars show the overlapping
-value). (C) Red bars show the overlapping  -value with CGC and GBM driver genes.
-value with CGC and GBM driver genes.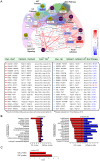
 -values between the pathway and OVC-related genes. (C) Red bars show the
-values between the pathway and OVC-related genes. (C) Red bars show the  -values that overlap with those of the CGC- and OVC-related genes.
-values that overlap with those of the CGC- and OVC-related genes.References
-
- Hahn WC, Weinberg RA (2002) Modelling the molecular circuitry of cancer. Nat Rev Cancer 2: 331–341. - PubMed
-
- Vogelstein B, Kinzler KW (2004) Cancer genes and the pathways they control. Nat Med 10: 789–799. - PubMed
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, et al. (2002) Mutations of the BRAF gene in human cancer. Nature 417: 949–954. - PubMed
-
- Wan PTC, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, et al. (2004) Mechanism of Activation of the RAF-ERK Signaling Pathway by Oncogenic Mutations of B-RAF. Cell 116: 855–867. - PubMed
-
- Santarosa M, Ashworth A (2004) Haploinsufficiency for tumour suppressor genes: when you don't need to go all the way. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 1654: 105–122. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

