Rapid global fitting of large fluorescence lifetime imaging microscopy datasets
- PMID: 23940626
- PMCID: PMC3734241
- DOI: 10.1371/journal.pone.0070687
Rapid global fitting of large fluorescence lifetime imaging microscopy datasets
Abstract
Fluorescence lifetime imaging (FLIM) is widely applied to obtain quantitative information from fluorescence signals, particularly using Förster Resonant Energy Transfer (FRET) measurements to map, for example, protein-protein interactions. Extracting FRET efficiencies or population fractions typically entails fitting data to complex fluorescence decay models but such experiments are frequently photon constrained, particularly for live cell or in vivo imaging, and this leads to unacceptable errors when analysing data on a pixel-wise basis. Lifetimes and population fractions may, however, be more robustly extracted using global analysis to simultaneously fit the fluorescence decay data of all pixels in an image or dataset to a multi-exponential model under the assumption that the lifetime components are invariant across the image (dataset). This approach is often considered to be prohibitively slow and/or computationally expensive but we present here a computationally efficient global analysis algorithm for the analysis of time-correlated single photon counting (TCSPC) or time-gated FLIM data based on variable projection. It makes efficient use of both computer processor and memory resources, requiring less than a minute to analyse time series and multiwell plate datasets with hundreds of FLIM images on standard personal computers. This lifetime analysis takes account of repetitive excitation, including fluorescence photons excited by earlier pulses contributing to the fit, and is able to accommodate time-varying backgrounds and instrument response functions. We demonstrate that this global approach allows us to readily fit time-resolved fluorescence data to complex models including a four-exponential model of a FRET system, for which the FRET efficiencies of the two species of a bi-exponential donor are linked, and polarisation-resolved lifetime data, where a fluorescence intensity and bi-exponential anisotropy decay model is applied to the analysis of live cell homo-FRET data. A software package implementing this algorithm, FLIMfit, is available under an open source licence through the Open Microscopy Environment.
Conflict of interest statement
Figures
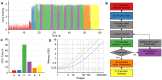
 frame time gated FLIM dataset, (red) a
frame time gated FLIM dataset, (red) a  TCSPC dataset and (green) a
TCSPC dataset and (green) a  two channel polarisation resolved dataset. Numbers exclude the memory required for the MATLAB runtime engine (300 MB).
two channel polarisation resolved dataset. Numbers exclude the memory required for the MATLAB runtime engine (300 MB).



References
-
- Vogel SS, Thaler C, Koushik SV (2006) Fanciful FRET. Science’s STKE 2006. doi:10.1126/stke.3312006re2. - PubMed
-
- Piston DW, Kremers GJ (2007) Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci 32: 407–414 doi:10.1016/j.tibs.2007.08.003 - DOI - PubMed
-
- Suhling K, Siegel J, Phillips D, French PMW, Lévêque-Fort S, et al. (2002) Imaging the environment of green fluorescent protein. Biophys J 83: 3589–3595 doi:10.1016/S0006-3495(02)75359-9 - DOI - PMC - PubMed
-
- Lakowicz JR (1999) Principles of Fluorescence Spectroscopy. Second Edi. Kluwer Academic.
-
- Munro I, McGinty J, Galletly N, Requejo-Isidro J, Lanigan PMP, et al. (2005) Toward the clinical application of time-domain fluorescence lifetime imaging. J Biomed Opt 10: 051403 doi:10.1117/1.2102807 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

