Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults
- PMID: 23940645
- PMCID: PMC3735522
- DOI: 10.1371/journal.pone.0070803
Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults
Abstract
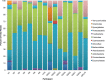
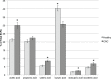
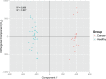
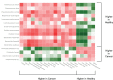
In this study we used stool profiling to identify intestinal bacteria and metabolites that are differentially represented in humans with colorectal cancer (CRC) compared to healthy controls to identify how microbial functions may influence CRC development. Stool samples were collected from healthy adults (n = 10) and colorectal cancer patients (n = 11) prior to colon resection surgery at the University of Colorado Health-Poudre Valley Hospital in Fort Collins, CO. The V4 region of the 16s rRNA gene was pyrosequenced and both short chain fatty acids and global stool metabolites were extracted and analyzed utilizing Gas Chromatography-Mass Spectrometry (GC-MS). There were no significant differences in the overall microbial community structure associated with the disease state, but several bacterial genera, particularly butyrate-producing species, were under-represented in the CRC samples, while a mucin-degrading species, Akkermansia muciniphila, was about 4-fold higher in CRC (p<0.01). Proportionately higher amounts of butyrate were seen in stool of healthy individuals while relative concentrations of acetate were higher in stools of CRC patients. GC-MS profiling revealed higher concentrations of amino acids in stool samples from CRC patients and higher poly and monounsaturated fatty acids and ursodeoxycholic acid, a conjugated bile acid in stool samples from healthy adults (p<0.01). Correlative analysis between the combined datasets revealed some potential relationships between stool metabolites and certain bacterial species. These associations could provide insight into microbial functions occurring in a cancer environment and will help direct future mechanistic studies. Using integrated "omics" approaches may prove a useful tool in identifying functional groups of gastrointestinal bacteria and their associated metabolites as novel therapeutic and chemopreventive targets.
Conflict of interest statement
Figures

References
-
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1131. - PubMed
-
- Kelly D, Conway S, Aminov R (2005) Commensal gut bacteria: mechanisms of immune modulation. Trends in Immunology 26: 326–333. - PubMed
-
- Klein RS, Catalano MT, Edberg SC, Casey JI, Steigbigel NH, et al. (1979) Streptococcus bovis septicemia and carcinoma of the colon. Ann Intern Med 91: 560–562. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

