Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range
- PMID: 23941402
- PMCID: PMC3751643
- DOI: 10.1186/1471-2164-14-551
Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range
Abstract
Background: Citrus bacterial canker is a disease that has severe economic impact on citrus industries worldwide and is caused by a few species and pathotypes of Xanthomonas. X. citri subsp. citri strain 306 (XccA306) is a type A (Asiatic) strain with a wide host range, whereas its variant X. citri subsp. citri strain A(w)12879 (Xcaw12879, Wellington strain) is restricted to Mexican lime.
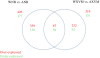
Results: To characterize the mechanism for the differences in host range of XccA and Xcaw, the genome of Xcaw12879 that was completed recently was compared with XccA306 genome. Effectors xopAF and avrGf1 are present in Xcaw12879, but were absent in XccA306. AvrGf1 was shown previously for Xcaw to cause hypersensitive response in Duncan grapefruit. Mutation analysis of xopAF indicates that the gene contributes to Xcaw growth in Mexican lime but does not contribute to the limited host range of Xcaw. RNA-Seq analysis was conducted to compare the expression profiles of Xcaw12879 and XccA306 in Nutrient Broth (NB) medium and XVM2 medium, which induces hrp gene expression. Two hundred ninety two and 281 genes showed differential expression in XVM2 compared to in NB for XccA306 and Xcaw12879, respectively. Twenty-five type 3 secretion system genes were up-regulated in XVM2 for both XccA and Xcaw. Among the 4,370 common genes of Xcaw12879 compared to XccA306, 603 genes in NB and 450 genes in XVM2 conditions were differentially regulated. Xcaw12879 showed higher protease activity than XccA306 whereas Xcaw12879 showed lower pectate lyase activity in comparison to XccA306.
Conclusions: Comparative genomic analysis of XccA306 and Xcaw12879 identified strain specific genes. Our study indicated that AvrGf1 contributes to the host range limitation of Xcaw12879 whereas XopAF contributes to virulence. Transcriptome analyses of XccA306 and Xcaw12879 presented insights into the expression of the two closely related strains of X. citri subsp. citri. Virulence genes including genes encoding T3SS components and effectors are induced in XVM2 medium. Numerous genes with differential expression in Xcaw12879 and XccA306 were identified. This study provided the foundation to further characterize the mechanisms for virulence and host range of pathotypes of X. citri subsp. citri.
Figures






Similar articles
-
Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host.Mol Plant Pathol. 2009 Mar;10(2):249-62. doi: 10.1111/j.1364-3703.2008.00528.x. Mol Plant Pathol. 2009. PMID: 19236573 Free PMC article.
-
New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library.BMC Microbiol. 2009 Jan 16;9:12. doi: 10.1186/1471-2180-9-12. BMC Microbiol. 2009. PMID: 19149882 Free PMC article.
-
High-throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development.Mol Plant Microbe Interact. 2012 Jan;25(1):69-84. doi: 10.1094/MPMI-05-11-0121. Mol Plant Microbe Interact. 2012. PMID: 21899385
-
Genetic and structural determinants on iron assimilation pathways in the plant pathogen Xanthomonas citri subsp. citri and Xanthomonas sp.Braz J Microbiol. 2020 Sep;51(3):1219-1231. doi: 10.1007/s42770-019-00207-x. Epub 2020 Aug 28. Braz J Microbiol. 2020. PMID: 31848911 Free PMC article. Review.
-
Xanthomonas as a Model System for Studying Pathogen Emergence and Evolution.Phytopathology. 2024 Jul;114(7):1433-1446. doi: 10.1094/PHYTO-03-24-0084-RVW. Epub 2024 Jul 5. Phytopathology. 2024. PMID: 38648116 Review.
Cited by
-
Stringent response regulators (p)ppGpp and DksA positively regulate virulence and host adaptation of Xanthomonas citri.Mol Plant Pathol. 2019 Nov;20(11):1550-1565. doi: 10.1111/mpp.12865. Epub 2019 Oct 17. Mol Plant Pathol. 2019. PMID: 31621195 Free PMC article.
-
Toxin-Antitoxin Gene Pairs Found in Tn3 Family Transposons Appear To Be an Integral Part of the Transposition Module.mBio. 2020 Mar 31;11(2):e00452-20. doi: 10.1128/mBio.00452-20. mBio. 2020. PMID: 32234815 Free PMC article.
-
Greater than the sum of their parts: an overview of the AvrRps4 effector family.Front Plant Sci. 2024 May 10;15:1400659. doi: 10.3389/fpls.2024.1400659. eCollection 2024. Front Plant Sci. 2024. PMID: 38799092 Free PMC article. Review.
-
Citrus Canker Pathogen, Its Mechanism of Infection, Eradication, and Impacts.Plants (Basel). 2022 Dec 26;12(1):123. doi: 10.3390/plants12010123. Plants (Basel). 2022. PMID: 36616252 Free PMC article. Review.
-
Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management.Mol Plant Pathol. 2018 Jun;19(6):1302-1318. doi: 10.1111/mpp.12638. Epub 2018 Mar 8. Mol Plant Pathol. 2018. PMID: 29105297 Free PMC article.
References
-
- Civerolo EL. Bacterial canker disease of citrus. J Rio Grande Val Hort Soc. 1984;37:127–145.
-
- Sun X, Stall RE, Jones JB, Cubero J, Gottwald TR, Graham JH, Dixon WH, Schubert TS, Chaloux PH, Stromberg VK. et al.Detection and characterization of a new strain of citrus canker bacteria from Key/Mexican Lime and alemow in south Florida. Plant Dis. 2004;88:1179–1188. doi: 10.1094/PDIS.2004.88.11.1179. - DOI - PubMed
-
- Verniere C, Hartung JS, Pruvost OP, Civerolo EL, Alvarez AM, Maestri P, Luisetti J. Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur J Plant Pathol. 1998;104:477–487. doi: 10.1023/A:1008676508688. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

