Therapeutic targeting of autophagy in disease: biology and pharmacology
- PMID: 23943849
- PMCID: PMC3799234
- DOI: 10.1124/pr.112.007120
Therapeutic targeting of autophagy in disease: biology and pharmacology
Abstract
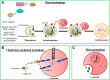
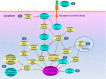
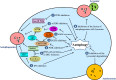
Autophagy, a process of self-digestion of the cytoplasm and organelles through which cellular components are recycled for reuse or energy production, is an evolutionarily conserved response to metabolic stress found in eukaryotes from yeast to mammals. It is noteworthy that autophagy is also associated with various pathophysiologic conditions in which this cellular process plays either a cytoprotective or cytopathic role in response to a variety of stresses such as metabolic, inflammatory, neurodegenerative, and therapeutic stress. It is now generally believed that modulating the activity of autophagy through targeting specific regulatory molecules in the autophagy machinery may impact disease processes, thus autophagy may represent a new pharmacologic target for drug development and therapeutic intervention of various human disorders. Induction or inhibition of autophagy using small molecule compounds has shown promise in the treatment of diseases such as cancer. Depending on context, induction or suppression of autophagy may exert therapeutic effects via promoting either cell survival or death, two major events targeted by therapies for various disorders. A better understanding of the biology of autophagy and the pharmacology of autophagy modulators has the potential for facilitating the development of autophagy-based therapeutic interventions for several human diseases.
Figures
References
-
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. (1999) Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 59:59–65 - PubMed
-
- Akazawa H, Komazaki S, Shimomura H, Terasaki F, Zou Y, Takano H, Nagai T, Komuro I. (2004) Diphtheria toxin-induced autophagic cardiomyocyte death plays a pathogenic role in mouse model of heart failure. J Biol Chem 279:41095–41103 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

