kClust: fast and sensitive clustering of large protein sequence databases
- PMID: 23945046
- PMCID: PMC3843501
- DOI: 10.1186/1471-2105-14-248
kClust: fast and sensitive clustering of large protein sequence databases
Abstract
Background: Fueled by rapid progress in high-throughput sequencing, the size of public sequence databases doubles every two years. Searching the ever larger and more redundant databases is getting increasingly inefficient. Clustering can help to organize sequences into homologous and functionally similar groups and can improve the speed, sensitivity, and readability of homology searches. However, because the clustering time is quadratic in the number of sequences, standard sequence search methods are becoming impracticable.
Results: Here we present a method to cluster large protein sequence databases such as UniProt within days down to 20%-30% maximum pairwise sequence identity. kClust owes its speed and sensitivity to an alignment-free prefilter that calculates the cumulative score of all similar 6-mers between pairs of sequences, and to a dynamic programming algorithm that operates on pairs of similar 4-mers. To increase sensitivity further, kClust can run in profile-sequence comparison mode, with profiles computed from the clusters of a previous kClust iteration. kClust is two to three orders of magnitude faster than clustering based on NCBI BLAST, and on multidomain sequences of 20%-30% maximum pairwise sequence identity it achieves comparable sensitivity and a lower false discovery rate. It also compares favorably to CD-HIT and UCLUST in terms of false discovery rate, sensitivity, and speed.
Conclusions: kClust fills the need for a fast, sensitive, and accurate tool to cluster large protein sequence databases to below 30% sequence identity. kClust is freely available under GPL at http://toolkit.lmb.uni-muenchen.de/pub/kClust/.
Figures


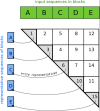



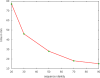

References
-
- Chubb D, Jefferys BR, Sternberg MJE, Kelley LA. Sequencing delivers diminishing returns for homology detection: implications for mapping the protein universe. Bioinformatics. 2010;26(21):2664–2671. [ http://bioinformatics.oxfordjournals.org/content/26/21/2664.abstract] - PubMed
-
- Li W, Jaroszewski L, Godzik A. Sequence clustering strategies improve remote homology recognitions while reducing search times. Protein Eng. 2002;15(8):643–649. [ http://view.ncbi.nlm.nih.gov/pubmed/12364578] - PubMed
-
- Park J, Holm L, Heger A, Chothia C. RSDB: representative protein sequence databases have high information content. Bioinformatics. 2000;16(5):458–464. [ http://view.ncbi.nlm.nih.gov/pubmed/10871268] - PubMed
-
- Suzek B, Huang H, McGarvey P, Mazumder R, Wu C. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23(10):1282–1288. - PubMed
-
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers Y, Falcón LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S. et al.The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5(3):e77. [ http://dx.doi.org/10.1371/journal.pbio.0050077] - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

