Differentiation and functional regulation of human fetal NK cells
- PMID: 23945237
- PMCID: PMC3754261
- DOI: 10.1172/JCI68989
Differentiation and functional regulation of human fetal NK cells
Abstract
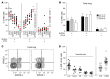
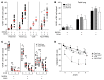
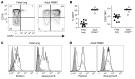
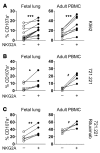
The human fetal immune system is naturally exposed to maternal allogeneic cells, maternal antibodies, and pathogens. As such, it is faced with a considerable challenge with respect to the balance between immune reactivity and tolerance. Here, we show that fetal natural killer (NK) cells differentiate early in utero and are highly responsive to cytokines and antibody-mediated stimulation but respond poorly to HLA class I-negative target cells. Strikingly, expression of killer-cell immunoglobulin-like receptors (KIRs) did not educate fetal NK cells but rendered them hyporesponsive to target cells lacking HLA class I. In addition, fetal NK cells were highly susceptible to TGF-β-mediated suppression, and blocking of TGF-β signaling enhanced fetal NK cell responses to target cells. Our data demonstrate that KIR-mediated hyporesponsiveness and TGF-β-mediated suppression are major factors determining human fetal NK cell hyporesponsiveness to HLA class I-negative target cells and provide a potential mechanism for fetal-maternal tolerance in utero. Finally, our results provide a basis for understanding the role of fetal NK cells in pregnancy complications in which NK cells could be involved, for example, during in utero infections and anti-RhD-induced fetal anemia.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

