Discovery of cell-type specific regulatory elements in the human genome using differential chromatin modification analysis
- PMID: 23945931
- PMCID: PMC3814353
- DOI: 10.1093/nar/gkt712
Discovery of cell-type specific regulatory elements in the human genome using differential chromatin modification analysis
Abstract
Chromatin modifications have been comprehensively illustrated to play important roles in gene regulation and cell diversity in recent years. Given the rapid accumulation of genome-wide chromatin modification maps across multiple cell types, there is an urgent need for computational methods to analyze multiple maps to reveal combinatorial modification patterns and define functional DNA elements, especially those are specific to cell types or tissues. In this current study, we developed a computational method using differential chromatin modification analysis (dCMA) to identify cell-type-specific genomic regions with distinctive chromatin modifications. We then apply this method to a public data set with modification profiles of nine marks for nine cell types to evaluate its effectiveness. We found cell-type-specific elements unique to each cell type investigated. These unique features show significant cell-type-specific biological relevance and tend to be located within functional regulatory elements. These results demonstrate the power of a differential comparative epigenomic strategy in deciphering the human genome and characterizing cell specificity.
Figures



 indicated by (asterisk). (D) CSRE neighboring genes are more likely to be non-housekeeping genes with the exception of those in K562 and GM12878. Dashed vertical line on the left side represent the P-value threshold (
indicated by (asterisk). (D) CSRE neighboring genes are more likely to be non-housekeeping genes with the exception of those in K562 and GM12878. Dashed vertical line on the left side represent the P-value threshold ( ). (E) CSRE neighboring genes show distinct functional enrichments highly relevant to the corresponding cell type contexts. We chose the top five enriched GO terms in each cell type, and
). (E) CSRE neighboring genes show distinct functional enrichments highly relevant to the corresponding cell type contexts. We chose the top five enriched GO terms in each cell type, and  was used to generate the heat map. (F) Enriched phenotypes of SNPs located in the CSREs in three cell types.
was used to generate the heat map. (F) Enriched phenotypes of SNPs located in the CSREs in three cell types.
 . The ‘CSRE neighboring’ gene group had higher expression levels, with exception of the H1 ES cell line. (B) Bar plot of the mean intensity of H3K27me3, a repressive chromatin modification, among all CSREs. (C) The comparison of transcription diversity of all CSRE neighboring genes and remaining genes. For each gene, the CV was used to measure the diversity of expression levels across the nine cell types examined. Boxplot comparison of the CV was shown, and the statistical significance was evaluated using two-sample Wilcoxon test.
. The ‘CSRE neighboring’ gene group had higher expression levels, with exception of the H1 ES cell line. (B) Bar plot of the mean intensity of H3K27me3, a repressive chromatin modification, among all CSREs. (C) The comparison of transcription diversity of all CSRE neighboring genes and remaining genes. For each gene, the CV was used to measure the diversity of expression levels across the nine cell types examined. Boxplot comparison of the CV was shown, and the statistical significance was evaluated using two-sample Wilcoxon test.
 e–16 for all cases.
e–16 for all cases.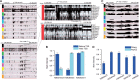
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

