FGF signaling regulates Wnt ligand expression to control vulval cell lineage polarity in C. elegans
- PMID: 23946444
- PMCID: PMC3754481
- DOI: 10.1242/dev.095687
FGF signaling regulates Wnt ligand expression to control vulval cell lineage polarity in C. elegans
Abstract
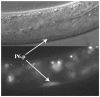
The interpretation of extracellular cues leading to the polarization of intracellular components and asymmetric cell divisions is a fundamental part of metazoan organogenesis. The Caenorhabditis elegans vulva, with its invariant cell lineage and interaction of multiple cell signaling pathways, provides an excellent model for the study of cell polarity within an organized epithelial tissue. Here, we show that the fibroblast growth factor (FGF) pathway acts in concert with the Frizzled homolog LIN-17 to influence the localization of SYS-1, a component of the Wnt/β-catenin asymmetry pathway, indirectly through the regulation of cwn-1. The source of the FGF ligand is the primary vulval precursor cell (VPC) P6.p, which controls the orientation of the neighboring secondary VPC P7.p by signaling through the sex myoblasts (SMs), activating the FGF pathway. The Wnt CWN-1 is expressed in the posterior body wall muscle of the worm as well as in the SMs, making it the only Wnt expressed on the posterior and anterior sides of P7.p at the time of the polarity decision. Both sources of cwn-1 act instructively to influence P7.p polarity in the direction of the highest Wnt signal. Using single molecule fluorescence in situ hybridization, we show that the FGF pathway regulates the expression of cwn-1 in the SMs. These results demonstrate an interaction between FGF and Wnt in C. elegans development and vulval cell lineage polarity, and highlight the promiscuous nature of Wnts and the importance of Wnt gradient directionality within C. elegans.
Keywords: C. elegans; Cell polarity; FGF; Vulval development; Wnt.
Figures



References
-
- Branda C. S., Stern M. J. (2000). Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol. 226, 137–151 - PubMed
-
- Burdine R. D., Branda C. S., Stern M. J. (1998). EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125, 1083–1093 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

