Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1
- PMID: 23946460
- PMCID: PMC3807354
- DOI: 10.1128/JVI.03433-12
Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1
Abstract
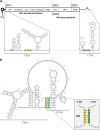
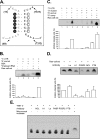
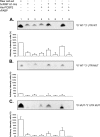
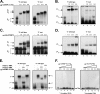
Sequences and structures within the terminal genomic regions of plus-strand RNA viruses are targets for the binding of host proteins that modulate functions such as translation, RNA replication, and encapsidation. Using murine norovirus 1 (MNV-1), we describe the presence of long-range RNA-RNA interactions that were stabilized by cellular proteins. The proteins potentially responsible for the stabilization were selected based on their ability to bind the MNV-1 genome and/or having been reported to be involved in the stabilization of RNA-RNA interactions. Cell extracts were preincubated with antibodies against the selected proteins and used for coprecipitation reactions. Extracts treated with antibodies to poly(C) binding protein 2 (PCBP2) and heterogeneous nuclear ribonucleoprotein (hnRNP) A1 significantly reduced the 5'-3' interaction. Both PCBP2 and hnRNP A1 recombinant proteins stabilized the 5'-3' interactions and formed ribonucleoprotein complexes with the 5' and 3' ends of the MNV-1 genomic RNA. Mutations within the 3' complementary sequences (CS) that disrupt the 5'-3'-end interactions resulted in a significant reduction of the viral titer, suggesting that the integrity of the 3'-end sequence and/or the lack of complementarity with the 5' end is important for efficient virus replication. Small interfering RNA-mediated knockdown of PCBP2 or hnRNP A1 resulted in a reduction in virus yield, confirming a role for the observed interactions in efficient viral replication. PCBP2 and hnRNP A1 induced the circularization of MNV-1 RNA, as revealed by electron microscopy. This study provides evidence that PCBP2 and hnRNP A1 bind to the 5' and 3' ends of the MNV-1 viral RNA and contribute to RNA circularization, playing a role in the virus life cycle.
Figures




Similar articles
-
Cellular protein hnRNP-A1 interacts with the 3'-end and the intergenic sequence of mouse hepatitis virus negative-strand RNA to form a ribonucleoprotein complex.Adv Exp Med Biol. 1998;440:227-34. doi: 10.1007/978-1-4615-5331-1_28. Adv Exp Med Biol. 1998. PMID: 9782285
-
Heterogeneous nuclear ribonucleoprotein a1 binds to the 3'-untranslated region and mediates potential 5'-3'-end cross talks of mouse hepatitis virus RNA.J Virol. 2001 Jun;75(11):5009-17. doi: 10.1128/JVI.75.11.5009-5017.2001. J Virol. 2001. PMID: 11333880 Free PMC article.
-
hnRNP L and NF90 interact with hepatitis C virus 5'-terminal untranslated RNA and promote efficient replication.J Virol. 2014 Jul;88(13):7199-209. doi: 10.1128/JVI.00225-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719423 Free PMC article.
-
Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles.Mol Cell Neurosci. 2013 Sep;56:436-46. doi: 10.1016/j.mcn.2012.12.002. Epub 2012 Dec 14. Mol Cell Neurosci. 2013. PMID: 23247072 Review.
-
A comprehensive understanding of hnRNP A1 role in cancer: new perspectives on binding with noncoding RNA.Cancer Gene Ther. 2023 Mar;30(3):394-403. doi: 10.1038/s41417-022-00571-1. Epub 2022 Dec 2. Cancer Gene Ther. 2023. PMID: 36460805 Review.
Cited by
-
A cytoplasmic quaking I isoform regulates the hnRNP F/H-dependent alternative splicing pathway in myelinating glia.Nucleic Acids Res. 2014 Jun;42(11):7319-29. doi: 10.1093/nar/gku353. Epub 2014 May 3. Nucleic Acids Res. 2014. PMID: 24792162 Free PMC article.
-
Production of infectious reporter murine norovirus by VP2 trans-complementation.J Virol. 2024 Feb 20;98(2):e0126123. doi: 10.1128/jvi.01261-23. Epub 2024 Jan 16. J Virol. 2024. PMID: 38226813 Free PMC article.
-
Noroviruses subvert the core stress granule component G3BP1 to promote viral VPg-dependent translation.Elife. 2019 Aug 12;8:e46681. doi: 10.7554/eLife.46681. Elife. 2019. PMID: 31403400 Free PMC article.
-
Binding of microRNA-122 to the hepatitis C virus 5' untranslated region modifies interactions with poly(C) binding protein 2 and the NS5B viral polymerase.Nucleic Acids Res. 2023 Dec 11;51(22):12397-12413. doi: 10.1093/nar/gkad1000. Nucleic Acids Res. 2023. PMID: 37941151 Free PMC article.
-
The circular RNA circ_GRHPR promotes NSCLC cell proliferation and invasion via interactions with the RNA-binding protein PCBP2.Clin Exp Pharmacol Physiol. 2021 Aug;48(8):1171-1181. doi: 10.1111/1440-1681.13523. Epub 2021 May 29. Clin Exp Pharmacol Physiol. 2021. PMID: 33987874 Free PMC article.
References
-
- Teunis PFM, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476 - PubMed
-
- Clarke IN, Lambden PR. 1997. The molecular biology of caliciviruses. J. Gen. Virol. 78:291–301 - PubMed
-
- McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, Heeney J, Yarovinsky F, Simmonds P, Macdonald A, Goodfellow I. 2011. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 7:e1002413.10.1371/journal.ppat.1002413 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

