Interaction of profilin with the barbed end of actin filaments
- PMID: 23947767
- PMCID: PMC3823579
- DOI: 10.1021/bi400682n
Interaction of profilin with the barbed end of actin filaments
Abstract
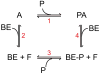
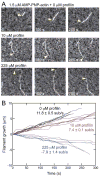
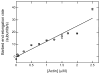
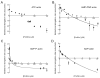
Profilin binds not only to actin monomers but also to the barbed end of the actin filament, where it inhibits association of subunits. To address open questions about the interactions of profilin with barbed ends, we measured the effects of a wide range of concentrations of Homo sapiens profilin 1 on the rate of elongation of individual skeletal muscle actin filaments by total internal reflection fluorescence microscopy. Much higher concentrations of profilin were required to stop elongation by AMP-PNP-actin monomers than ADP-actin monomers. High concentrations of profilin depolymerized barbed ends at a rate much faster than the spontaneous dissociation rates of Mg-ATP-, Mg-AMP-PNP-, Mg-ADP-Pi-, and Mg-ADP-actin subunits. Fitting a thermodynamic model to these data allowed us to determine the affinities of profilin and profilin-actin for barbed ends and the influence of the nucleotide bound to actin on these interactions. Profilin has a much higher affinity for ADP-actin filament barbed ends (Kd = 1 μM) than AMP-PNP-actin filament barbed ends (Kd = 226 μM). ADP-actin monomers associated with profilin bind to ADP-actin filament barbed ends 10% as fast as free ADP-actin monomers, but bound profilin does not affect the rate of association of AMP-PNP-actin monomers with barbed ends. The differences in the affinities of AMP-PNP- and ADP-bound barbed ends for profilin and profilin-actin suggest that conformations of barbed end subunits differ from those of monomers and change upon nucleotide hydrolysis and phosphate release. A structural model revealed minor steric clashes between profilin and actin subunits at the barbed end that explain the biochemical results.
Figures

References
-
- Birbach A. Profilin, a multi-modal regulator of neuronal plasticity. Bioessays. 2008;30:994–1002. - PubMed
-
- Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. - PubMed
-
- Markey F, Larsson H, Weber K, Lindberg U. Nucleation of actin polymerization from profilactin opposite effects of different nuclei. Biochim Biophys Acta. 1982;704:43–51. - PubMed
-
- Tobacman LS, Brenner SL, Korn ED. Effect of Acanthamoeba profilin on the pre-steady state kinetics of actin polymerization and on the oncentration of F-actin at steady state. J Biol Chem. 1983;258:8806–8812. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

