New and TALENted genome engineering toolbox
- PMID: 23948583
- PMCID: PMC3965580
- DOI: 10.1161/CIRCRESAHA.113.301765
New and TALENted genome engineering toolbox
Abstract
Recent advances in the burgeoning field of genome engineering are accelerating the realization of personalized therapeutics for cardiovascular disease. In the postgenomic era, sequence-specific gene-editing tools enable the functional analysis of genetic alterations implicated in disease. In partnership with high-throughput model systems, efficient gene manipulation provides an increasingly powerful toolkit to study phenotypes associated with patient-specific genetic defects. Herein, this review emphasizes the latest developments in genome engineering and how applications within the field are transforming our understanding of personalized medicine with an emphasis on cardiovascular diseases.
Keywords: TALENs; cardiovascular disease modeling; genome engineering; induced pluripotent stem cells; zebrafish.
Figures

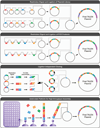
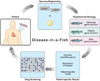
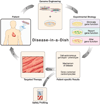
References
-
- Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart Disease and Stroke Statistics—2013 Update: A Report From the American Heart Association. Circulation. 2013;127:e6–e245. - PMC - PubMed
-
- Maron BJ, Maron MS, Semsarian C. Genetics of Hypertrophic Cardiomyopathy After 20 Years: Clinical Perspectives. J Am Coll Cardiol. 2012;60:705–715. - PubMed
-
- Kimura A. Molecular Etiology and Pathogenesis of Hereditary Cardiomyopathy. Circ J. 2008;72:A38–A48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

