Role of DNMT3A, TET2, and IDH1/2 mutations in pre-leukemic stem cells in acute myeloid leukemia
- PMID: 23949914
- PMCID: PMC5542003
- DOI: 10.1007/s12185-013-1407-8
Role of DNMT3A, TET2, and IDH1/2 mutations in pre-leukemic stem cells in acute myeloid leukemia
Abstract
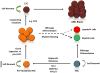
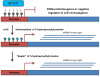
Aberrant changes in the epigenome are now recognized to be important in driving the development of multiple human cancers including acute myeloid leukemia. Recent advances in sequencing technologies have led to the identification of recurrent mutations in genes that regulate DNA methylation including DNA methyltransferase 3A (DNMT3A), ten-eleven translocation 2 (TET2), and isocitrate dehydrogenase 1 (IDH1) and IDH2. These mutations have been shown to promote self-renewal and block differentiation of hematopoietic stem/progenitor cells. Acquisition of these mutations in hematopoietic stem cells can lead to their clonal expansion resulting in a pre-leukemic stem cell (pre-LSC) population. Pre-LSCs retain the ability to differentiate into the full spectrum of mature daughter cells but can become fully transformed with the acquisition of additional driver mutations. Here, we review the effects of mutations in DNMT3A, TET2, and IDH1/2 on mouse and human hematopoiesis, the current understanding of their role in pre-LSCs, and therapeutic strategies to eliminate this population which may serve as a cellular reservoir for relapse.
Figures


References
-
- Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nature immunology. 2004;5:738–43. - PubMed
-
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nature reviews Cancer. 2012;12:599–612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

