Female behaviour drives expression and evolution of gustatory receptors in butterflies
- PMID: 23950722
- PMCID: PMC3732137
- DOI: 10.1371/journal.pgen.1003620
Female behaviour drives expression and evolution of gustatory receptors in butterflies
Abstract
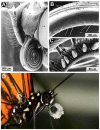
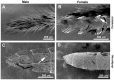
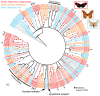
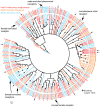
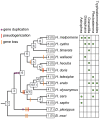
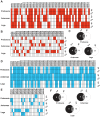
Secondary plant compounds are strong deterrents of insect oviposition and feeding, but may also be attractants for specialist herbivores. These insect-plant interactions are mediated by insect gustatory receptors (Grs) and olfactory receptors (Ors). An analysis of the reference genome of the butterfly Heliconius melpomene, which feeds on passion-flower vines (Passiflora spp.), together with whole-genome sequencing within the species and across the Heliconius phylogeny has permitted an unprecedented opportunity to study the patterns of gene duplication and copy-number variation (CNV) among these key sensory genes. We report in silico gene predictions of 73 Gr genes in the H. melpomene reference genome, including putative CO2, sugar, sugar alcohol, fructose, and bitter receptors. The majority of these Grs are the result of gene duplications since Heliconius shared a common ancestor with the monarch butterfly or the silkmoth. Among Grs but not Ors, CNVs are more common within species in those gene lineages that have also duplicated over this evolutionary time-scale, suggesting ongoing rapid gene family evolution. Deep sequencing (∼1 billion reads) of transcriptomes from proboscis and labial palps, antennae, and legs of adult H. melpomene males and females indicates that 67 of the predicted 73 Gr genes and 67 of the 70 predicted Or genes are expressed in these three tissues. Intriguingly, we find that one-third of all Grs show female-biased gene expression (n = 26) and nearly all of these (n = 21) are Heliconius-specific Grs. In fact, a significant excess of Grs that are expressed in female legs but not male legs are the result of recent gene duplication. This difference in Gr gene expression diversity between the sexes is accompanied by a striking sexual dimorphism in the abundance of gustatory sensilla on the forelegs of H. melpomene, suggesting that female oviposition behaviour drives the evolution of new gustatory receptors in butterfly genomes.
Conflict of interest statement
The authors have declared that no competing interests exit.
Figures




References
-
- Ehrlich PR, Raven PH (1964) Butterflies and plants: A study in coevolution. Evolution 18: 586–608.
-
- Dethier VG (1937) Gustation and olfaction in lepidopterous larvæ. Biol Bull 72: 7–23.
-
- Schneider D (1964) Insect antennae. Annu Rev Entomol 9: 103–122.
-
- Schoonhoven LM, Dethier VM (1966) Sensory aspects of host-plant discrimination by lepidopterous larvae. Arch Neerl ZooI 16: 497–530.
-
- Anderson AL (1932) The sensitivity of the legs of common butterflies to sugars. J Exp Zool B Mol Dev Evol 63: 235–259.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

