Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells
- PMID: 23951130
- PMCID: PMC3737158
- DOI: 10.1371/journal.pone.0071316
Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells
Abstract
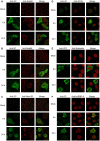
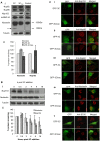
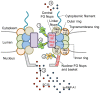
Human Rhinovirus (HRV) infection results in shut down of essential cellular processes, in part through disruption of nucleocytoplasmic transport by cleavage of the nucleoporin proteins (Nups) that make up the host cell nuclear pore. Although the HRV genome encodes two proteases (2A and 3C) able to cleave host proteins such as Nup62, little is known regarding the specific contribution of each. Here we use transfected as well as HRV-infected cells to establish for the first time that 3C protease is most likely the mediator of cleavage of Nup153 during HRV infection, while Nup62 and Nup98 are likely to be targets of HRV2A protease. HRV16 3C protease was also able to elicit changes in the appearance and distribution of the nuclear speckle protein SC35 in transfected cells, implicating it as a key mediator of the mislocalisation of SC35 in HRV16-infected cells. In addition, 3C protease activity led to the redistribution of the nucleolin protein out of the nucleolus, but did not affect nuclear localisation of hnRNP proteins, implying that complete disruption of nucleocytoplasmic transport leading to relocalisation of hnRNP proteins from the nucleus to the cytoplasm in HRV-infected cells almost certainly requires 2A in addition to 3C protease. Thus, a specific role for HRV 3C protease in cleavage and mislocalisation of host cell nuclear proteins, in concert with 2A, is implicated for the first time in HRV pathogenesis.
Conflict of interest statement
Figures

References
-
- Racaniello VR (2001) Picornaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology, 4th Edition. Philadelphia, PA: Lippincott/Raven. 685–722.
-
- Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K (2011) Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411: 374–382. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

