IFN-ε is constitutively expressed by cells of the reproductive tract and is inefficiently secreted by fibroblasts and cell lines
- PMID: 23951133
- PMCID: PMC3739789
- DOI: 10.1371/journal.pone.0071320
IFN-ε is constitutively expressed by cells of the reproductive tract and is inefficiently secreted by fibroblasts and cell lines
Abstract
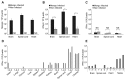
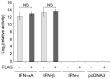
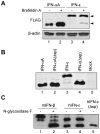
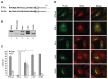
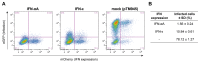
Type-I interferons (IFNs) form a large family of cytokines that primarily act to control the early development of viral infections. Typical type-I IFN genes, such as those encoding IFN-α or IFN-β are upregulated by viral infection in many cell types. In contrast, the gene encoding IFN-ε was reported to be constitutively expressed by cells of the female reproductive tract and to contribute to the protection against vaginal infections with herpes simplex virus 2 and Chlamydia muridarum. Our data confirm the lack of induction of IFN-ε expression after viral infection and the constitutive expression of IFN-ε by cells of the female but also of the male reproductive organs. Interestingly, when expressed from transfected expression plasmids in 293T, HeLa or Neuro2A cells, the mouse and human IFN-ε precursors were inefficiently processed and secretion of IFN-ε was minimal. Analysis of chimeric constructs produced between IFN-ε and limitin (IFN-ζ) showed that both the signal peptide and the mature moiety of IFN-ε contribute to poor processing of the precursor. Immunofluorescent detection of FLAG-tagged IFN-ε in transfected cells suggested that IFN-ε and chimeric proteins were defective for progression through the secretory pathway. IFN-ε did not, however, act intracellularly and impart an antiviral state to producing cells. Given the constitutive expression of IFN-ε in specialized cells and the poor processing of IFN-ε precursor in fibroblasts and cell lines, we hypothesize that IFN-ε secretion may require a co-factor specifically expressed in cells of the reproductive organs, that might secure the system against aberrant release of this IFN.
Conflict of interest statement
Figures

References
-
- Isaacs A, Lindenmann J (1957) Virus interference. I. Interferon Proc R Soc Lond B Biol Sci 147: 258-267. doi:10.1098/rspb.1957.0048. - DOI - PubMed
-
- Uzé G, Schreiber G, Piehler J, Pellegrini S (2007) The receptor of the type I interferon family. Curr Top Microbiol Immunol 316: 71-95. doi:10.1007/978-3-540-71329-6_5. PubMed: 17969444. - DOI - PubMed
-
- Liu SY, Sanchez DJ, Cheng G (2011) New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol 23: 57-64. doi:10.1016/j.coi.2010.11.003. PubMed: 21123041. - DOI - PMC - PubMed
-
- Randall RE, Goodbourn S (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89: 1-47. doi:10.1099/vir.0.83391-0. PubMed: 18089727. - DOI - PubMed
-
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT et al. (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472: 481-485. doi:10.1038/nature09907. PubMed: 21478870. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

