Comparison of molecular breeding values based on within- and across-breed training in beef cattle
- PMID: 23953034
- PMCID: PMC3765540
- DOI: 10.1186/1297-9686-45-30
Comparison of molecular breeding values based on within- and across-breed training in beef cattle
Abstract
Background: Although the efficacy of genomic predictors based on within-breed training looks promising, it is necessary to develop and evaluate across-breed predictors for the technology to be fully applied in the beef industry. The efficacies of genomic predictors trained in one breed and utilized to predict genetic merit in differing breeds based on simulation studies have been reported, as have the efficacies of predictors trained using data from multiple breeds to predict the genetic merit of purebreds. However, comparable studies using beef cattle field data have not been reported.
Methods: Molecular breeding values for weaning and yearling weight were derived and evaluated using a database containing BovineSNP50 genotypes for 7294 animals from 13 breeds in the training set and 2277 animals from seven breeds (Angus, Red Angus, Hereford, Charolais, Gelbvieh, Limousin, and Simmental) in the evaluation set. Six single-breed and four across-breed genomic predictors were trained using pooled data from purebred animals. Molecular breeding values were evaluated using field data, including genotypes for 2227 animals and phenotypic records of animals born in 2008 or later. Accuracies of molecular breeding values were estimated based on the genetic correlation between the molecular breeding value and trait phenotype.
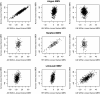
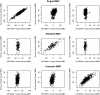
Results: With one exception, the estimated genetic correlations of within-breed molecular breeding values with trait phenotype were greater than 0.28 when evaluated in the breed used for training. Most estimated genetic correlations for the across-breed trained molecular breeding values were moderate (> 0.30). When molecular breeding values were evaluated in breeds that were not in the training set, estimated genetic correlations clustered around zero.
Conclusions: Even for closely related breeds, within- or across-breed trained molecular breeding values have limited prediction accuracy for breeds that were not in the training set. For breeds in the training set, across- and within-breed trained molecular breeding values had similar accuracies. The benefit of adding data from other breeds to a within-breed training population is the ability to produce molecular breeding values that are more robust across breeds and these can be utilized until enough training data has been accumulated to allow for a within-breed training set.
Figures


References
-
- Thallman RM, Hanford KJ, Quaas RL, Kachman SD, Tempelman RJ, Fernando RL, Kuehn LA, Pollak EJ. Estimation of the proportion of genetic variation accounted for by DNA tests. Proceedings of the Beef Improvement Federation 41st Annual Research Symposium and Annual Meeting: April 30-May 3 2009: Sacramento. 2009. pp. 184–209.
-
- MacNeil MD, Northcutt SL, Schnabel RD, Garrick DJ, Woodward BW, Taylor JF. Genetic correlations between carcass traits and molecular breeding values in Angus cattle. Proceedings of the 9th World Congress Applied to Livestock Production: 1–6 August 2010, Leipzig. 2010. pp. 2–148.
-
- Pfizer Animal Health and American Brahman Breeders Association Introduce Tenderness GE-EPD. 2012. [ http://beefmagazine.com/selection-tools/pfizer-animal-health-and-america...-]
-
- Johnston D, Graser H, Tier B. Research and development of trial Brahman BREEDPLAN Tenderness EBVM. 2008. [ http://agbu.une.edu.au/brahman%20tenderness%20EBVs.pdf]
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

