HIV latency and integration site placement in five cell-based models
- PMID: 23953889
- PMCID: PMC3765678
- DOI: 10.1186/1742-4690-10-90
HIV latency and integration site placement in five cell-based models
Abstract
Background: HIV infection can be treated effectively with antiretroviral agents, but the persistence of a latent reservoir of integrated proviruses prevents eradication of HIV from infected individuals. The chromosomal environment of integrated proviruses has been proposed to influence HIV latency, but the determinants of transcriptional repression have not been fully clarified, and it is unclear whether the same molecular mechanisms drive latency in different cell culture models.
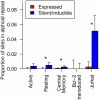
Results: Here we compare data from five different in vitro models of latency based on primary human T cells or a T cell line. Cells were infected in vitro and separated into fractions containing proviruses that were either expressed or silent/inducible, and integration site populations sequenced from each. We compared the locations of 6,252 expressed proviruses to those of 6,184 silent/inducible proviruses with respect to 140 forms of genomic annotation, many analyzed over chromosomal intervals of multiple lengths. A regularized logistic regression model linking proviral expression status to genomic features revealed no predictors of latency that performed better than chance, though several genomic features were significantly associated with proviral expression in individual models. Proviruses in the same chromosomal region did tend to share the same expressed or silent/inducible status if they were from the same cell culture model, but not if they were from different models.
Conclusions: The silent/inducible phenotype appears to be associated with chromosomal position, but the molecular basis is not fully clarified and may differ among in vitro models of latency.
Figures







References
-
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. http://www.nature.com/nm/journal/v1/n12/full/nm1295-1284.html. - DOI - PubMed
-
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. http://dx.doi.org/10.1038/387183a0. - DOI - DOI - PubMed
-
- Davey RT, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96(26):15109–15114. doi: 10.1073/pnas.96.26.15109. http://dx.doi.org/10.1073/pnas.96.26.15109. - DOI - DOI - PMC - PubMed
-
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323(5919):1304–1307. doi: 10.1126/science.1165706. http://dx.doi.org/10.1126/science.1165706. - DOI - DOI - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. http://dx.doi.org/10.1038/8394. - DOI - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- R21AI 096993/AI/NIAID NIH HHS/United States
- AI087508/AI/NIAID NIH HHS/United States
- 5T32HG000046/HG/NHGRI NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- R01 AI052845/AI/NIAID NIH HHS/United States
- T32 AI007384/AI/NIAID NIH HHS/United States
- K02AI078766/AI/NIAID NIH HHS/United States
- R01 AI 052845-11/AI/NIAID NIH HHS/United States
- U19 AI096113/AI/NIAID NIH HHS/United States
- U19AI096113/AI/NIAID NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
- P30 AI 045008/AI/NIAID NIH HHS/United States
- R01AI038201/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

