Plasticity in the contribution of T cell receptor variable region residues to binding of peptide-HLA-A2 complexes
- PMID: 23954306
- PMCID: PMC3844621
- DOI: 10.1016/j.jmb.2013.08.007
Plasticity in the contribution of T cell receptor variable region residues to binding of peptide-HLA-A2 complexes
Abstract
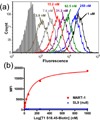
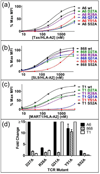
One hypothesis accounting for major histocompatibility complex (MHC) restriction by T cell receptors (TCRs) holds that there are several evolutionary conserved residues in TCR variable regions that contact MHC. While this "germline codon" hypothesis is supported by various lines of evidence, it has been difficult to test. The difficulty stems in part from the fact that TCRs exhibit low affinities for pep/MHC, thus limiting the range of binding energies that can be assigned to these key interactions using mutational analyses. To measure the magnitude of binding energies involved, here we used high-affinity TCRs engineered by mutagenesis of CDR3. The TCRs included a high-affinity, MART-1/HLA-A2-specific single-chain TCR and two other high-affinity TCRs that all contain the same Vα region and recognize the same MHC allele (HLA-A2), with different peptides and Vβ regions. Mutational analysis of residues in CDR1 and CDR2 of the three Vα2 regions showed the importance of the key germline codon residue Y51. However, two other proposed key residues showed significant differences among the TCRs in their relative contributions to binding. With the use of single-position, yeast-display libraries in two of the key residues, MART-1/HLA-A2 selections also revealed strong preferences for wild-type germline codon residues, but several alternative residues could also accommodate binding and, hence, MHC restriction. Thus, although a single residue (Y51) could account for a proportion of the energy associated with positive selection (i.e., MHC restriction), there is significant plasticity in requirements for particular side chains in CDR1 and CDR2 and in their relative binding contributions among different TCRs.
Keywords: CDR; MHC; SOE; SPR; T cell receptor; T cell receptors; TCR; complementarity-determining region; directed evolution; germline codon bias; major histocompatibility complex; single-chain; splicing by overlap extension; surface plasmon resonance; yeast display.
© 2013.
Figures




Similar articles
-
Subtle changes at the variable domain interface of the T-cell receptor can strongly increase affinity.J Biol Chem. 2018 Feb 2;293(5):1820-1834. doi: 10.1074/jbc.M117.814152. Epub 2017 Dec 11. J Biol Chem. 2018. PMID: 29229779 Free PMC article.
-
T-cell receptor (TCR)-peptide specificity overrides affinity-enhancing TCR-major histocompatibility complex interactions.J Biol Chem. 2014 Jan 10;289(2):628-38. doi: 10.1074/jbc.M113.522110. Epub 2013 Nov 6. J Biol Chem. 2014. PMID: 24196962 Free PMC article.
-
Structural basis for ineffective T-cell responses to MHC anchor residue-improved "heteroclitic" peptides.Eur J Immunol. 2015 Feb;45(2):584-91. doi: 10.1002/eji.201445114. Epub 2014 Dec 28. Eur J Immunol. 2015. PMID: 25471691 Free PMC article.
-
Evolutionarily conserved amino acids that control TCR-MHC interaction.Annu Rev Immunol. 2008;26:171-203. doi: 10.1146/annurev.immunol.26.021607.090421. Annu Rev Immunol. 2008. PMID: 18304006 Free PMC article. Review.
-
T cells and their eons-old obsession with MHC.Immunol Rev. 2012 Nov;250(1):49-60. doi: 10.1111/imr.12004. Immunol Rev. 2012. PMID: 23046122 Free PMC article. Review.
Cited by
-
An Engineered Switch in T Cell Receptor Specificity Leads to an Unusual but Functional Binding Geometry.Structure. 2016 Jul 6;24(7):1142-1154. doi: 10.1016/j.str.2016.04.011. Epub 2016 May 26. Structure. 2016. PMID: 27238970 Free PMC article.
-
Molecular properties of gp100-reactive T-cell receptors drive the cytokine profile and antitumor efficacy of transgenic host T cells.Pigment Cell Melanoma Res. 2019 Jan;32(1):68-78. doi: 10.1111/pcmr.12724. Epub 2018 Aug 13. Pigment Cell Melanoma Res. 2019. PMID: 30009548 Free PMC article.
-
Comparison of T Cell Activities Mediated by Human TCRs and CARs That Use the Same Recognition Domains.J Immunol. 2018 Feb 1;200(3):1088-1100. doi: 10.4049/jimmunol.1700236. Epub 2017 Dec 29. J Immunol. 2018. PMID: 29288199 Free PMC article.
-
A novel T cell receptor single-chain signaling complex mediates antigen-specific T cell activity and tumor control.Cancer Immunol Immunother. 2014 Nov;63(11):1163-76. doi: 10.1007/s00262-014-1586-z. Epub 2014 Aug 1. Cancer Immunol Immunother. 2014. PMID: 25082071 Free PMC article.
-
Changing the peptide specificity of a human T-cell receptor by directed evolution.Nat Commun. 2014 Nov 7;5:5223. doi: 10.1038/ncomms6223. Nat Commun. 2014. PMID: 25376839 Free PMC article.
References
-
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. - PubMed
-
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

