Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism
- PMID: 23954414
- PMCID: PMC4231430
- DOI: 10.1016/j.cell.2013.07.035
Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism
Abstract
The molecular mechanism of autophagy and its relationship to other lysosomal degradation pathways remain incompletely understood. Here, we identified a previously uncharacterized mammalian-specific protein, Beclin 2, which, like Beclin 1, functions in autophagy and interacts with class III PI3K complex components and Bcl-2. However, Beclin 2, but not Beclin 1, functions in an additional lysosomal degradation pathway. Beclin 2 is required for ligand-induced endolysosomal degradation of several G protein-coupled receptors (GPCRs) through its interaction with GASP1. Beclin 2 homozygous knockout mice have decreased embryonic viability, and heterozygous knockout mice have defective autophagy, increased levels of brain cannabinoid 1 receptor, elevated food intake, and obesity and insulin resistance. Our findings identify Beclin 2 as a converging regulator of autophagy and GPCR turnover and highlight the functional and mechanistic diversity of Beclin family members in autophagy, endolysosomal trafficking, and metabolism.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

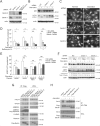
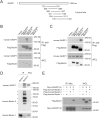
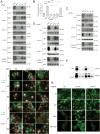

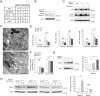

Comment in
-
Common and divergent functions of Beclin 1 and Beclin 2.Cell Res. 2013 Dec;23(12):1341-2. doi: 10.1038/cr.2013.129. Epub 2013 Sep 10. Cell Res. 2013. PMID: 24018378 Free PMC article.
Similar articles
-
Endolysosomal trafficking of viral G protein-coupled receptor functions in innate immunity and control of viral oncogenesis.Proc Natl Acad Sci U S A. 2016 Mar 15;113(11):2994-9. doi: 10.1073/pnas.1601860113. Epub 2016 Feb 29. Proc Natl Acad Sci U S A. 2016. PMID: 26929373 Free PMC article.
-
Differential interactions between Beclin 1 and Bcl-2 family members.Autophagy. 2007 Nov-Dec;3(6):561-8. doi: 10.4161/auto.4713. Epub 2007 Jul 8. Autophagy. 2007. PMID: 17643073
-
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
-
The potent BECN2-ATG14 coiled-coil interaction is selectively critical for endolysosomal degradation of GPRASP1/GASP1-associated GPCRs.Autophagy. 2023 Nov;19(11):2884-2898. doi: 10.1080/15548627.2023.2233872. Epub 2023 Jul 13. Autophagy. 2023. PMID: 37409929 Free PMC article.
-
The Beclin 1 network regulates autophagy and apoptosis.Cell Death Differ. 2011 Apr;18(4):571-80. doi: 10.1038/cdd.2010.191. Epub 2011 Feb 11. Cell Death Differ. 2011. PMID: 21311563 Free PMC article. Review.
Cited by
-
Autophagy mediates tolerance to Staphylococcus aureus alpha-toxin.Cell Host Microbe. 2015 Apr 8;17(4):429-40. doi: 10.1016/j.chom.2015.03.001. Epub 2015 Mar 26. Cell Host Microbe. 2015. PMID: 25816775 Free PMC article.
-
Broad targeting of resistance to apoptosis in cancer.Semin Cancer Biol. 2015 Dec;35 Suppl(0):S78-S103. doi: 10.1016/j.semcancer.2015.03.001. Epub 2015 Apr 28. Semin Cancer Biol. 2015. PMID: 25936818 Free PMC article. Review.
-
Sesamin-mediated high expression of BECN2 ameliorates cartilage endplate degeneration by reducing autophagy and inflammation.Aging (Albany NY). 2024 Jan 26;16(2):1145-1160. doi: 10.18632/aging.205386. Epub 2024 Jan 26. Aging (Albany NY). 2024. PMID: 38284902 Free PMC article.
-
BECN1/Beclin 1 sorts cell-surface APP/amyloid β precursor protein for lysosomal degradation.Autophagy. 2016 Dec;12(12):2404-2419. doi: 10.1080/15548627.2016.1234561. Epub 2016 Oct 7. Autophagy. 2016. PMID: 27715386 Free PMC article.
-
Exocytosis Proteins: Typical and Atypical Mechanisms of Action in Skeletal Muscle.Front Endocrinol (Lausanne). 2022 Jun 14;13:915509. doi: 10.3389/fendo.2022.915509. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35774142 Free PMC article. Review.
References
-
- Abu-Helo A, Simonin F. Identification and biological significance of G protein-coupled receptor associated sorting proteins (GASPs) Pharmacol Ther. 2010;126:244–250. - PubMed
-
- Aissani B, Perusse L, Lapointe G, Chagnon YC, Bouchard L, Walts B, Bouchard C. A quantitative trait locus for body fat on chromosome 1q43 in French Canadians: linkage and association studies. Obesity (Silver Spring) 2006;14:1605–1615. - PubMed
-
- Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Perusse L, Vohl MC. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J Lipid Res. 2004;45:419–426. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- P01 DK088761/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R21 AI078108/AI/NIAID NIH HHS/United States
- P20 RR015566/RR/NCRR NIH HHS/United States
- R01 DA019958/DA/NIDA NIH HHS/United States
- R0I CA109618/CA/NCI NIH HHS/United States
- R01 GM094575/GM/NIGMS NIH HHS/United States
- K99 DK094980/DK/NIDDK NIH HHS/United States
- R01 CA109618/CA/NCI NIH HHS/United States
- R0I DA019958/DA/NIDA NIH HHS/United States
- R0I DK55758/DK/NIDDK NIH HHS/United States
- P30 GM103332/GM/NIGMS NIH HHS/United States
- R21 AI078198/AI/NIAID NIH HHS/United States
- R0I DK043351/DK/NIDDK NIH HHS/United States
- R0I GM094575/GM/NIGMS NIH HHS/United States
- R01 DK055758/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

