Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response
- PMID: 23954590
- PMCID: PMC4196939
- DOI: 10.1016/j.devcel.2013.07.015
Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response
Abstract
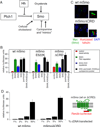
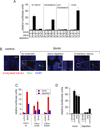
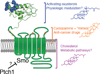
Hedgehog (Hh) signaling during development and in postembryonic tissues requires activation of the 7TM oncoprotein Smoothened (Smo) by mechanisms that may involve endogenous lipidic modulators. Exogenous Smo ligands previously identified include the plant sterol cyclopamine (and its therapeutically useful synthetic mimics) and hydroxylated cholesterol derivatives (oxysterols); Smo is also highly sensitive to cellular sterol levels. The relationships between these effects are unclear because the relevant Smo structural determinants are unknown. We identify the conserved extracellular cysteine-rich domain (CRD) as the site of action for oxysterols on Smo, involving residues structurally analogous to those contacting the Wnt lipid adduct in the homologous Frizzled CRD; this modulatory effect is distinct from that of cyclopamine mimics, from Hh-mediated regulation, and from the permissive action of cellular sterol pools. These results imply that Hh pathway activity is sensitive to lipid binding at several Smo sites, suggesting mechanisms for tuning by multiple physiological inputs.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Ayers KL, Thérond PP. Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends in Cell Biology. 2010;20:287–298. - PubMed
-
- Bazan JF, de Sauvage FJ. Structural ties between cholesterol transport and morphogen signaling. Cell. 2009;138:1055–1056. - PubMed
-
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

