The epidermis comprises autonomous compartments maintained by distinct stem cell populations
- PMID: 23954751
- PMCID: PMC3793873
- DOI: 10.1016/j.stem.2013.07.010
The epidermis comprises autonomous compartments maintained by distinct stem cell populations
Abstract
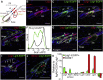
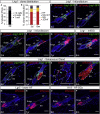
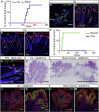
The complex anatomy of the epidermis contains multiple adult stem cell populations, but the extent to which they functionally overlap during homeostasis, wound healing, and tumor initiation remains poorly defined. Here, we demonstrate that Lrig1(+ve) cells are highly proliferative epidermal stem cells. Long-term clonal analysis reveals that Lrig1(+ve) cells maintain the upper pilosebaceous unit, containing the infundibulum and sebaceous gland as independent compartments, but contribute to neither the hair follicle nor the interfollicular epidermis, which are maintained by distinct stem cell populations. In contrast, upon wounding, stem cell progeny from multiple compartments acquire lineage plasticity and make permanent contributions to regenerating tissue. We further show that oncogene activation in Lrig1(+ve) cells drives hyperplasia but requires auxiliary stimuli for tumor formation. In summary, our data demonstrate that epidermal stem cells are lineage restricted during homeostasis and suggest that compartmentalization may constitute a conserved mechanism underlying epithelial tissue maintenance.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Alcolea M.P., Jones P.H. Tracking cells in their native habitat: lineage tracing in epithelial neoplasia. Nat. Rev. Cancer. 2013;13:161–171. - PubMed
-
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Barker N., van Oudenaarden A., Clevers H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell. 2012;11:452–460. - PubMed
-
- Blanpain C. Stem cells: Skin regeneration and repair. Nature. 2010;464:686–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

