Nanoparticle for delivery of antisense γPNA oligomers targeting CCR5
- PMID: 23954968
- PMCID: PMC3771998
- DOI: 10.4161/adna.25628
Nanoparticle for delivery of antisense γPNA oligomers targeting CCR5
Abstract
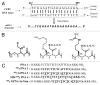
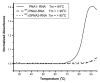
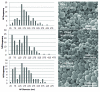
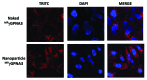
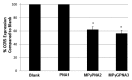
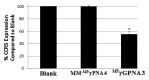
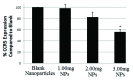
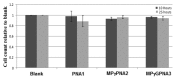
The development of a new class of peptide nucleic acids (PNAs), i.e., gamma PNAs (γPNAs), creates the need for a general and effective method for its delivery into cells for regulating gene expression in mammalian cells. Here we report the antisense activity of a recently developed hydrophilic and biocompatible diethylene glycol (miniPEG)-based gamma peptide nucleic acid called MPγPNAs via its delivery by poly(lactide-co-glycolide) (PLGA)-based nanoparticle system. We show that MPγPNA oligomers designed to bind to the selective region of chemokine receptor 5 (CC R5) transcript, induce potent and sequence-specific antisense effects as compared with regular PNA oligomers. In addition, PLGA nanoparticle delivery of MPγPNAs is not toxic to the cells. The findings reported in this study provide a combination of γPNA technology and PLGA-based nanoparticle delivery method for regulating gene expression in live cells via the antisense mechanism.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
