Acrylyl-coenzyme A reductase, an enzyme involved in the assimilation of 3-hydroxypropionate by Rhodobacter sphaeroides
- PMID: 23955006
- PMCID: PMC3807440
- DOI: 10.1128/JB.00685-13
Acrylyl-coenzyme A reductase, an enzyme involved in the assimilation of 3-hydroxypropionate by Rhodobacter sphaeroides
Abstract
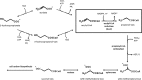
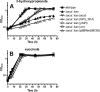
The anoxygenic phototroph Rhodobacter sphaeroides uses 3-hydroxypropionate as a sole carbon source for growth. Previously, we showed that the gene (RSP_1434) known as acuI, which encodes a protein of the medium-chain dehydrogenase/reductase (MDR) superfamily, was involved in 3-hydroxypropionate assimilation via the reductive conversion to propionyl-coenzyme A (CoA). Based on these results, we speculated that acuI encoded acrylyl-CoA reductase. In this work, we characterize the in vitro enzyme activity of purified, recombinant AcuI using a coupled spectrophotometric assay. AcuI from R. sphaeroides catalyzes the NADPH-dependent acrylyl-CoA reduction to produce propionyl-CoA. Two other members of the MDR012 family within the MDR superfamily, the products of SPO_1914 from Ruegeria pomeroyi and yhdH from Escherichia coli, were shown to also be part of this new class of NADPH-dependent acrylyl-CoA reductases. The activities of the three enzymes were characterized by an extremely low Km for acrylyl-CoA (<3 μM) and turnover numbers of 45 to 80 s(-1). These homodimeric enzymes were highly specific for NADPH (Km = 18 to 33 μM), with catalytic efficiencies of more than 10-fold higher for NADPH than for NADH. The introduction of codon-optimized SPO_1914 or yhdH into a ΔacuI::kan mutant of R. sphaeroides on a plasmid complemented 3-hydroxypropionate-dependent growth. However, in their native hosts, SPO_1914 and yhdH are believed to function in the metabolism of substrates other than 3-hydroxypropionate, where acrylyl-CoA is an intermediate. Complementation of the ΔacuI::kan mutant phenotype by crotonyl-CoA carboxylase/reductase from R. sphaeroides was attributed to the fact that the enzyme also uses acrylyl-CoA as a substrate.
Figures


References
-
- Alber BE, Fuchs G. 2002. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 277:12137–12143 - PubMed
-
- Hetzel M, Brock M, Selmer T, Pierik AJ, Golding BT, Buckel W. 2003. Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur. J. Biochem. 270:902–910 - PubMed
-
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in archaea. Science 318:1782–1786 - PubMed
-
- Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE, Fuchs G. 2010. Autotrophic carbon fixation in archaea. Nat. Rev. Microbiol. 8:447–460 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

