Bacterial colonization factors control specificity and stability of the gut microbiota
- PMID: 23955152
- PMCID: PMC3893107
- DOI: 10.1038/nature12447
Bacterial colonization factors control specificity and stability of the gut microbiota
Abstract
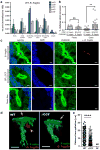
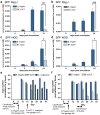
Mammals harbour a complex gut microbiome, comprising bacteria that confer immunological, metabolic and neurological benefits. Despite advances in sequence-based microbial profiling and myriad studies defining microbiome composition during health and disease, little is known about the molecular processes used by symbiotic bacteria to stably colonize the gastrointestinal tract. We sought to define how mammals assemble and maintain the Bacteroides, one of the most numerically prominent genera of the human microbiome. Here we find that, whereas the gut normally contains hundreds of bacterial species, germ-free mice mono-associated with a single Bacteroides species are resistant to colonization by the same, but not different, species. To identify bacterial mechanisms for species-specific saturable colonization, we devised an in vivo genetic screen and discovered a unique class of polysaccharide utilization loci that is conserved among intestinal Bacteroides. We named this genetic locus the commensal colonization factors (ccf). Deletion of the ccf genes in the model symbiont, Bacteroides fragilis, results in colonization defects in mice and reduced horizontal transmission. The ccf genes of B. fragilis are upregulated during gut colonization, preferentially at the colonic surface. When we visualize microbial biogeography within the colon, B. fragilis penetrates the colonic mucus and resides deep within crypt channels, whereas ccf mutants are defective in crypt association. Notably, the CCF system is required for B. fragilis colonization following microbiome disruption with Citrobacter rodentium infection or antibiotic treatment, suggesting that the niche within colonic crypts represents a reservoir for bacteria to maintain long-term colonization. These findings reveal that intestinal Bacteroides have evolved species-specific physical interactions with the host that mediate stable and resilient gut colonization, and the CCF system represents a novel molecular mechanism for symbiosis.
Figures