RNase III is required for actinomycin production in Streptomyces antibioticus
- PMID: 23956389
- PMCID: PMC3811219
- DOI: 10.1128/AEM.02272-13
RNase III is required for actinomycin production in Streptomyces antibioticus
Abstract
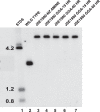
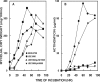
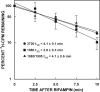
Using insertional mutagenesis, we have disrupted the RNase III gene, rnc, of the actinomycin-producing streptomycete, Streptomyces antibioticus. Disruption was verified by Southern blotting. The resulting strain grows more vigorously than its parent on actinomycin production medium but produces significantly lower levels of actinomycin. Complementation of the rnc disruption with the wild-type rnc gene from S. antibioticus restored actinomycin production to nearly wild-type levels. Western blotting experiments demonstrated that the disruptant did not produce full-length or truncated forms of RNase III. Thus, as is the case in Streptomyces coelicolor, RNase III is required for antibiotic production in S. antibioticus. No differences in the chemical half-lives of bulk mRNA were observed in a comparison of the S. antibioticus rnc mutant and its parental strain.
Figures
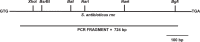

References
-
- Drider D, Condon C. 2004. The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 8:195–200 - PubMed
-
- Ji X. 2006. Structural basis for non-catalytic and catalytic activities of ribonuclease III. Acta Crystallogr. D 62:933–940 - PubMed
-
- Nicholson AW. 1999. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23:371–390 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

