Identifying potential RNAi targets in grain aphid (Sitobion avenae F.) based on transcriptome profiling of its alimentary canal after feeding on wheat plants
- PMID: 23957588
- PMCID: PMC3751716
- DOI: 10.1186/1471-2164-14-560
Identifying potential RNAi targets in grain aphid (Sitobion avenae F.) based on transcriptome profiling of its alimentary canal after feeding on wheat plants
Abstract
Background: The grain aphid (Sitobion avenae F.) is a major agricultural pest which causes significant yield losses of wheat in China, Europe and North America annually. Transcriptome profiling of the grain aphid alimentary canal after feeding on wheat plants could provide comprehensive gene expression information involved in feeding, ingestion and digestion. Furthermore, selection of aphid-specific RNAi target genes would be essential for utilizing a plant-mediated RNAi strategy to control aphids via a non-toxic mode of action. However, due to the tiny size of the alimentary canal and lack of genomic information on grain aphid as a whole, selection of the RNAi targets is a challenging task that as far as we are aware, has never been documented previously.
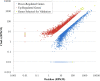
Results: In this study, we performed de novo transcriptome assembly and gene expression analyses of the alimentary canals of grain aphids before and after feeding on wheat plants using Illumina RNA sequencing. The transcriptome profiling generated 30,427 unigenes with an average length of 664 bp. Furthermore, comparison of the transcriptomes of alimentary canals of pre- and post feeding grain aphids indicated that 5490 unigenes were differentially expressed, among which, diverse genes and/or pathways were identified and annotated. Based on the RPKM values of these unigenes, 16 of them that were significantly up or down-regulated upon feeding were selected for dsRNA artificial feeding assay. Of these, 5 unigenes led to higher mortality and developmental stunting in an artificial feeding assay due to the down-regulation of the target gene expression. Finally, by adding fluorescently labelled dsRNA into the artificial diet, the spread of fluorescence signal in the whole body tissues of grain aphid was observed.
Conclusions: Comparison of the transcriptome profiles of the alimentary canals of pre- and post-feeding grain aphids on wheat plants provided comprehensive gene expression information that could facilitate our understanding of the molecular mechanisms underlying feeding, ingestion and digestion. Furthermore, five novel and effective potential RNAi target genes were identified in grain aphid for the first time. This finding would provide a fundamental basis for aphid control in wheat through plant mediated RNAi strategy.
Figures

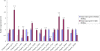

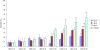
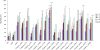


Similar articles
-
RNA Interference of the Ecdysone Receptor Genes EcR and USP in Grain Aphid (Sitobion avenae F.) Affects Its Survival and Fecundity upon Feeding on Wheat Plants.Int J Mol Sci. 2016 Dec 14;17(12):2098. doi: 10.3390/ijms17122098. Int J Mol Sci. 2016. PMID: 27983619 Free PMC article.
-
Double-stranded RNA in the biological control of grain aphid (Sitobion avenae F.).Funct Integr Genomics. 2015 Mar;15(2):211-23. doi: 10.1007/s10142-014-0424-x. Epub 2014 Dec 3. Funct Integr Genomics. 2015. PMID: 25467938
-
Silencing an aphid-specific gene SmDSR33 for aphid control through plant-mediated RNAi in wheat.Front Plant Sci. 2023 Jan 9;13:1100394. doi: 10.3389/fpls.2022.1100394. eCollection 2022. Front Plant Sci. 2023. PMID: 36699834 Free PMC article.
-
Comparative transcriptome and histological analyses of wheat in response to phytotoxic aphid Schizaphis graminum and non-phytotoxic aphid Sitobion avenae feeding.BMC Plant Biol. 2019 Dec 10;19(1):547. doi: 10.1186/s12870-019-2148-5. BMC Plant Biol. 2019. PMID: 31823722 Free PMC article.
-
RNAi-mediated plant protection against aphids.Pest Manag Sci. 2016 Jun;72(6):1090-8. doi: 10.1002/ps.4258. Epub 2016 Mar 21. Pest Manag Sci. 2016. PMID: 26888776 Review.
Cited by
-
Silencing an essential gene involved in infestation and digestion in grain aphid through plant-mediated RNA interference generates aphid-resistant wheat plants.Plant Biotechnol J. 2019 May;17(5):852-854. doi: 10.1111/pbi.13067. Epub 2019 Feb 6. Plant Biotechnol J. 2019. PMID: 30582665 Free PMC article. No abstract available.
-
RNA Interference for Improving Disease Resistance in Plants and Its Relevance in This Clustered Regularly Interspaced Short Palindromic Repeats-Dominated Era in Terms of dsRNA-Based Biopesticides.Front Plant Sci. 2022 May 13;13:885128. doi: 10.3389/fpls.2022.885128. eCollection 2022. Front Plant Sci. 2022. PMID: 35645997 Free PMC article. Review.
-
Sequencing and validation of reference genes to analyze endogenous gene expression and quantify yellow dwarf viruses using RT-qPCR in viruliferous Rhopalosiphum padi.PLoS One. 2014 May 8;9(5):e97038. doi: 10.1371/journal.pone.0097038. eCollection 2014. PLoS One. 2014. PMID: 24810421 Free PMC article.
-
Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt).Sci Rep. 2016 May 11;6:25883. doi: 10.1038/srep25883. Sci Rep. 2016. PMID: 27165720 Free PMC article.
-
Addressing the challenges of symbiont-mediated RNAi in aphids.PeerJ. 2023 Feb 28;11:e14961. doi: 10.7717/peerj.14961. eCollection 2023. PeerJ. 2023. PMID: 36874963 Free PMC article.
References
-
- Blackman RL, Eastop VF. Aphids on the world's crops. An identification and information guide. Hoboken, New Jersey: Wiley, John & Sons, Incorporated; 1984. p. 476.
-
- Oerke EC. In: Crop production and crop protection: estimated losses in major food and cash crops. Oerke EC, Dehne HW, Schonbeck F, Weber A, editor. Amsterdam: Elsevier; 1994. Estimated crop losses in wheat; pp. 179–296.
-
- Morrison WP, Peairs FB. Response model concept and economic impact. Response model for an introduced pest-the Russian wheat aphid. Lanham, MD: Entomological Society of America; 1998.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

