The genome in three dimensions: a new frontier in human brain research
- PMID: 23958183
- PMCID: PMC3925763
- DOI: 10.1016/j.biopsych.2013.07.015
The genome in three dimensions: a new frontier in human brain research
Abstract
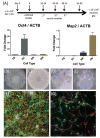
Less than 1.5% of the human genome encodes protein. However, vast portions of the human genome are subject to transcriptional and epigenetic regulation, and many noncoding regulatory DNA elements are thought to regulate the spatial organization of interphase chromosomes. For example, chromosomal "loopings" are pivotal for the orderly process of gene expression, by enabling distal regulatory enhancer or silencer elements to directly interact with proximal promoter and transcription start sites, potentially bypassing hundreds of kilobases of interspersed sequence on the linear genome. To date, however, epigenetic studies in the human brain are mostly limited to the exploration of DNA methylation and posttranslational modifications of the nucleosome core histones. In contrast, very little is known about the regulation of supranucleosomal structures. Here, we show that chromosome conformation capture, a widely used approach to study higher-order chromatin, is applicable to tissue collected postmortem, thereby informing about genome organization in the human brain. We introduce chromosome conformation capture protocols for brain and compare higher-order chromatin structures at the chromosome 6p22.2-22.1 schizophrenia and bipolar disorder susceptibility locus, and additional neurodevelopmental risk genes, (DPP10, MCPH1) in adult prefrontal cortex and various cell culture systems, including neurons derived from reprogrammed skin cells. We predict that the exploration of three-dimensional genome architectures and function will open up new frontiers in human brain research and psychiatric genetics and provide novel insights into the epigenetic risk architectures of regulatory noncoding DNA.
Keywords: Chromatin fiber; chromosomal looping; chromosome conformation capture; genome in 3D; higher-order chromatin; human brain.
Copyright © 2014 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Authors report no biomedical financial interests or potential conflicts of interest.
Figures



References
-
- Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. Journal of medical genetics. 2011;48:721–730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

