Early innate immunity determines outcome of Mycobacterium tuberculosis pulmonary infection in rabbits
- PMID: 23958185
- PMCID: PMC3765177
- DOI: 10.1186/1478-811X-11-60
Early innate immunity determines outcome of Mycobacterium tuberculosis pulmonary infection in rabbits
Abstract
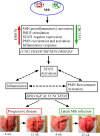
Background: Pulmonary infection of humans by Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), results in active disease in 5-10% of individuals, while asymptomatic latent Mtb infection (LTBI) is established in the remainder. The host immune responses that determine this differential outcome following Mtb infection are not fully understood. Using a rabbit model of pulmonary TB, we have shown that infection with the Mtb clinical isolate HN878 (a hyper-virulent W-Beijing lineage strain) leads to progressive cavitary disease similar to what is seen in humans with active TB. In contrast, infection with Mtb CDC1551 (a hyper-immunogenic clinical isolate) is efficiently controlled in rabbit lungs, with establishment of LTBI, which can be reactivated upon treatment with immune-suppressive drugs. We hypothesize that the initial interaction of Mtb with the cells of the host response in the lungs determine later outcome of infection.
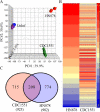
Results: To test this hypothesis, we used our rabbit model of pulmonary TB and infected the animals with Mtb HN878 or CDC1551. At 3 hours, with similar lung bacillary loads, HN878 infection caused greater accumulation of mononuclear and polymorphonuclear leukocytes (PMN) in the lungs, compared to animals infected with CDC1551. Using whole-genome microarray gene expression analysis, we delineated the early transcriptional changes in the lungs of HN878- or CDC1551-infected rabbits at this time and compared them to the differential response at 4 weeks of Mtb-infection. Our gene network and pathway analysis showed that the most significantly differentially expressed genes involved in the host response to HN878, compared to CDC1551, at 3 hours of infection, were components of the inflammatory response and STAT1 activation, recruitment and activation of macrophages, PMN, and fMLP (N-formyl-Methionyl-Leucyl-Phenylalanine)-stimulation. At 4 weeks, the CDC1551 bacillary load was significantly lower and the granulomatous response reduced compared to HN878 infection. Moreover, although inflammation was dampened in both Mtb infections at 4 weeks, the majority of the differentially expressed gene networks were similar to those seen at 3 hours.
Conclusions: We propose that differential regulation of the inflammation-associated innate immune response and related gene expression changes seen at 3 hours determine the long term outcome of Mtb infection in rabbit lungs.
Figures





References
-
- WHO. WHO report: Tuberculosis Fact Sheet 2008. Geneva: World Health Organization; 2008.
-
- Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol. 2012;12(5):352–366. - PubMed
-
- Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, Lalvani A, Wright D, Latif M, Davidson RN. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case–control study. Lancet. 2000;355(9204):618–621. doi: 10.1016/S0140-6736(99)02301-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

