Characterization of three novel adhesins of Leptospira interrogans
- PMID: 23958908
- PMCID: PMC3854887
- DOI: 10.4269/ajtmh.13-0205
Characterization of three novel adhesins of Leptospira interrogans
Abstract
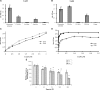
We report cloning, expression, purification, and characterization of three predicted leptospiral membrane proteins (LIC11360, LIC11009, and LIC11975). In silico analysis and proteinase K accessibility data suggest that these proteins might be surface exposed. We show that proteins encoded by LIC11360, LIC11009 and LIC11975 genes interact with laminin in a dose-dependent and saturable manner. The proteins are referred to as leptospiral surface adhesions 23, 26, and 36 (Lsa23, Lsa26, and Lsa36), respectively. These proteins also bind plasminogen and generate active plasmin. Attachment of Lsa23 and Lsa36 to fibronectin occurs through the involvement of the 30-kDa and 70-kDa heparin-binding domains of the ligand. Dose-dependent, specific-binding of Lsa23 to the complement regulator C4BP and to a lesser extent, to factor H, suggests that this protein may interfere with the complement cascade pathways. Leptospira spp. may use these interactions as possible mechanisms during the establishment of infection.
Figures







Similar articles
-
Features of two proteins of Leptospira interrogans with potential role in host-pathogen interactions.BMC Microbiol. 2012 Mar 30;12:50. doi: 10.1186/1471-2180-12-50. BMC Microbiol. 2012. PMID: 22463075 Free PMC article.
-
Lsa30, a novel adhesin of Leptospira interrogans binds human plasminogen and the complement regulator C4bp.Microb Pathog. 2012 Sep;53(3-4):125-34. doi: 10.1016/j.micpath.2012.06.001. Epub 2012 Jun 23. Microb Pathog. 2012. PMID: 22732096
-
Leptospira interrogans Lsa23 protein recruits plasminogen, factor H and C4BP from normal human serum and mediates C3b and C4b degradation.Microbiology (Reading). 2016 Feb;162(2):295-308. doi: 10.1099/mic.0.000217. Epub 2015 Nov 27. Microbiology (Reading). 2016. PMID: 26614523
-
A Review on Host-Leptospira Interactions: What We Know and Future Expectations.Front Cell Infect Microbiol. 2021 Nov 25;11:777709. doi: 10.3389/fcimb.2021.777709. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34900757 Free PMC article. Review.
-
Pathogenesis of leptospirosis: the influence of genomics.Vet Microbiol. 2011 Nov 21;153(1-2):73-81. doi: 10.1016/j.vetmic.2011.02.055. Epub 2011 Mar 5. Vet Microbiol. 2011. PMID: 21440384 Review.
Cited by
-
Features of two new proteins with OmpA-like domains identified in the genome sequences of Leptospira interrogans.PLoS One. 2015 Apr 7;10(4):e0122762. doi: 10.1371/journal.pone.0122762. eCollection 2015. PLoS One. 2015. PMID: 25849456 Free PMC article.
-
Leptospiral adhesins: from identification to future perspectives.Front Microbiol. 2024 Aug 13;15:1458655. doi: 10.3389/fmicb.2024.1458655. eCollection 2024. Front Microbiol. 2024. PMID: 39206373 Free PMC article. Review.
-
Unveiling the impact of Leptospira TolC efflux protein on host tissue adherence, complement evasion, and diagnostic potential.Infect Immun. 2024 Nov 12;92(11):e0041924. doi: 10.1128/iai.00419-24. Epub 2024 Oct 11. Infect Immun. 2024. PMID: 39392312 Free PMC article.
-
Evaluation of Leptospira interrogans knockdown mutants for LipL32, LipL41, LipL21, and OmpL1 proteins.Front Microbiol. 2023 Jun 23;14:1199660. doi: 10.3389/fmicb.2023.1199660. eCollection 2023. Front Microbiol. 2023. PMID: 37426019 Free PMC article.
-
Adhesins of Leptospira interrogans mediate the interaction to fibrinogen and inhibit fibrin clot formation in vitro.PLoS Negl Trop Dis. 2013 Aug 29;7(8):e2396. doi: 10.1371/journal.pntd.0002396. eCollection 2013. PLoS Negl Trop Dis. 2013. PMID: 24009788 Free PMC article.
References
-
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. - PubMed
-
- de la Pena-Moctezuma A, Bulach DM, Adler B Genetic differences among the LPS biosynthetic loci of serovars of Leptospira interrogans and Leptospira borgpetersenii. FEMS Immunol Med Microbiol. 2001;31:73–81. - PubMed
-
- Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. Melbourne. Australia: MediSci Press; 1999.
-
- Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

