The non-structural (NS) gene segment of H9N2 influenza virus isolated from backyard poultry in Pakistan reveals strong genetic and functional similarities to the NS gene of highly pathogenic H5N1
- PMID: 23959028
- PMCID: PMC3906295
- DOI: 10.4161/viru.26055
The non-structural (NS) gene segment of H9N2 influenza virus isolated from backyard poultry in Pakistan reveals strong genetic and functional similarities to the NS gene of highly pathogenic H5N1
Abstract
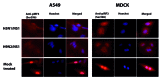
Apart from natural reassortment, co-circulation of different avian influenza virus strains in poultry populations can lead to generation of novel variants and reassortant viruses. In this report, we studied the genetics and functions of a reassorted non-structural gene (NS) of H9N2 influenza virus collected from back yard poultry (BYP) flock. Phylogenetic reconstruction based on hemagglutinin and neuraminidase genes indicates that an isolate from BYP belongs to H9N2. However, the NS gene-segment of this isolate cluster into genotype Z, clade 2.2 of the highly pathogenic H5N1. The NS gene plays essential roles in the host-adaptation, cell-tropism, and virulence of influenza viruses. However, such interpretations have not been investigated in naturally recombinant H9N2 viruses. Therefore, we compared the NS1 protein of H9N2 (H9N2/NS1) and highly pathogenic H5N1 (H5N1/NS1) in parallel for their abilities to regulate different signaling pathways, and investigated the molecular mechanisms of IFN-β production in human, avian, and mink lung cells. We found that H9N2/NS1 and H5N1/NS1 are comparably similar in inhibiting TNF-α induced nuclear factor κB and double stranded RNA induced activator protein 1 and interferon regulatory factor 3 transcription factors. Thus, the production of IFN-β was inhibited equally by both NS1s as demonstrated by IFN stimulatory response element and IFN-β promoter activation. Moreover, both NS1s predominantly localized in the nucleus when transfected to human A549 cells. This study therefore suggests the possible increased virulence of natural reassortant viruses for their efficient invasion of host immune responses, and proposes that these should not be overlooked for their epizootic and zoonotic potential.
Keywords: IFN production; NS1; immune regulation; influenza; reassortment.
Figures





Comment in
-
Avian influenza virus NS1: A small protein with diverse and versatile functions.Virulence. 2013 Oct 1;4(7):583-8. doi: 10.4161/viru.26360. Epub 2013 Sep 17. Virulence. 2013. PMID: 24051601 Free PMC article. No abstract available.
References
-
- Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A. 2000;97:9654–8. doi: 10.1073/pnas.160270697. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
