Molecular mechanisms of CRISPR-mediated microbial immunity
- PMID: 23959171
- PMCID: PMC3890593
- DOI: 10.1007/s00018-013-1438-6
Molecular mechanisms of CRISPR-mediated microbial immunity
Abstract
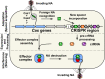
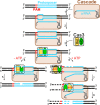
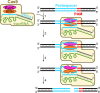
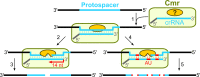
Bacteriophages (phages) infect bacteria in order to replicate and burst out of the host, killing the cell, when reproduction is completed. Thus, from a bacterial perspective, phages pose a persistent lethal threat to bacterial populations. Not surprisingly, bacteria evolved multiple defense barriers to interfere with nearly every step of phage life cycles. Phages respond to this selection pressure by counter-evolving their genomes to evade bacterial resistance. The antagonistic interaction between bacteria and rapidly diversifying viruses promotes the evolution and dissemination of bacteriophage-resistance mechanisms in bacteria. Recently, an adaptive microbial immune system, named clustered regularly interspaced short palindromic repeats (CRISPR) and which provides acquired immunity against viruses and plasmids, has been identified. Unlike the restriction–modification anti-phage barrier that subjects to cleavage any foreign DNA lacking a protective methyl-tag in the target site, the CRISPR–Cas systems are invader-specific, adaptive, and heritable. In this review, we focus on the molecular mechanisms of interference/immunity provided by different CRISPR–Cas systems.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

