Treatment with suboptimal doses of raltegravir leads to aberrant HIV-1 integrations
- PMID: 23959861
- PMCID: PMC3767498
- DOI: 10.1073/pnas.1305066110
Treatment with suboptimal doses of raltegravir leads to aberrant HIV-1 integrations
Abstract
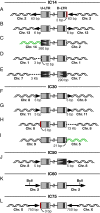
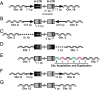
Integration of the DNA copy of the HIV-1 genome into a host chromosome is required for viral replication and is thus an important target for antiviral therapy. The HIV-encoded enzyme integrase (IN) catalyzes two essential steps: 3' processing of the viral DNA ends, followed by the strand transfer reaction, which inserts the viral DNA into host DNA. Raltegravir binds to IN and blocks the integration of the viral DNA. Using the Rous sarcoma virus-derived vector RCAS, we previously showed that mutations that cause one viral DNA end to be defective for IN-mediated integration led to abnormal integrations in which the provirus had one normal and one aberrant end, accompanied by rearrangements in the host genome. On the basis of these results, we expected that suboptimal concentrations of IN inhibitors, which could block one of the ends of viral integration, would lead to similar aberrant integrations. In contrast to the proviruses from untreated cells, which were all normal, ∼10-15% of the proviruses isolated after treatment with a suboptimal dose of raltegravir were aberrant. The aberrant integrations were similar to those seen in the RCAS experiments. Most of the aberrant proviruses had one normal end and one aberrant end and were accompanied by significant rearrangements in the host genome, including duplications, inversions, deletions and, occasionally, acquisition of sequences from other chromosomes. The rearrangements of the host DNA raise concerns that these aberrant integrations might have unintended consequences in HIV-1-infected patients who are not consistent in following a raltegravir-containing treatment regimen.
Keywords: chromosomal rearrangements; integrase inhibitors; strand transfer inhibitor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
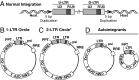
References
-
- Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol. 2011;21(2):249–256. - PubMed
-
- Coffin JM, Hughes SH, Varmus HE. The interactions of retroviruses and their hosts. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

