Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring
- PMID: 23959936
- PMCID: PMC3800750
- DOI: 10.1210/en.2012-2255
Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring
Abstract
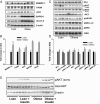
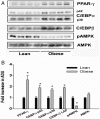
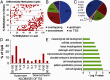
The risk of obesity (OB) in adulthood is strongly influenced by maternal body composition. Here we examined the hypothesis that maternal OB influences white adipose tissue (WAT) transcriptome and increases propensity for adipogenesis in the offspring, prior to the development of OB, using an established model of long-term metabolic programming. Employing an overfeeding-based rat model, in which exposure to OB is limited to preconception and gestation alone, we conducted global transcriptomic profiling in WAT, and gene/protein expression analysis of lipogenic and adipogenic pathways and examined adipogenic differentiation of WAT stromal-vascular cells ex vivo. Using reduced representation bisulfite sequencing we also evaluated genome-scale changes in DNA methylation in offspring WAT. Maternal OB led to extensive changes in expression of genes (± 1.8-fold, P ≤ .05), revealing a distinct up-regulation of lipogenic pathways in WAT. mRNA expression of a battery of sterol regulatory element-binding protein-1-regulated genes was increased in OB-dam offspring, which were confirmed by immunoblotting. In conjunction with lipogenic gene expression, OB-dam offspring showed increased glucose transporter-4 mRNA/protein expression and greater AKT phosphorylation following acute insulin challenge, suggesting sensitization of insulin signaling in WAT. Offspring of OB dams also exhibited increased in vivo expression of adipogenic regulators (peroxisome proliferator-activated receptor-γ, CCAAT enhancer binding protein α [C/EBP-α] and C/EBP-β), associated with greater ex vivo differentiation of WAT stromal-vascular cells. These transcriptomic changes were associated with alterations in DNA methylation of CpG sites and CGI shores, proximal to developmentally important genes, including key pro-adipogenic factors (Zfp423 and C/EBP-β). Our findings strongly suggest that the maternal OB in utero alters adipocyte commitment and differentiation via epigenetic mechanisms.
Figures





References
-
- Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA. 2012;307:491–497 - PubMed
-
- Nathanielsz PW, Poston L, Taylor PD. In utero exposure to maternal obesity and diabetes: animal models that identify and characterize implications for future health. Clin Perinatol. 2007;34:515–526, v - PubMed
-
- Shankar K, Harrell A, Liu X, Gilchrist JM, Ronis MJ, Badger TM. Maternal obesity at conception programs obesity in the offspring. Am J Physiol Regul Integr Comp Physiol. 2008;294:R528–R538 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

