The N-degradome of Escherichia coli: limited proteolysis in vivo generates a large pool of proteins bearing N-degrons
- PMID: 23960079
- PMCID: PMC3789986
- DOI: 10.1074/jbc.M113.492108
The N-degradome of Escherichia coli: limited proteolysis in vivo generates a large pool of proteins bearing N-degrons
Abstract
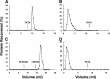
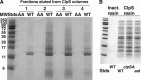
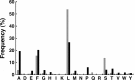
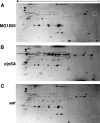
The N-end rule is a conserved mechanism found in Gram-negative bacteria and eukaryotes for marking proteins to be degraded by ATP-dependent proteases. Specific N-terminal amino acids (N-degrons) are sufficient to target a protein to the degradation machinery. In Escherichia coli, the adaptor ClpS binds an N-degron and delivers the protein to ClpAP for degradation. As ClpS recognizes N-terminal Phe, Trp, Tyr, and Leu, which are not found at the N terminus of proteins translated and processed by the canonical pathway, proteins must be post-translationally modified to expose an N-degron. One modification is catalyzed by Aat, an enzyme that adds leucine or phenylalanine to proteins with N-terminal lysine or arginine; however, such proteins are also not generated by the canonical protein synthesis pathway. Thus, the mechanisms producing N-degrons in proteins and the frequency of their occurrence largely remain a mystery. To address these issues, we used a ClpS affinity column to isolate interacting proteins from E. coli cell lysates under non-denaturing conditions. We identified more than 100 proteins that differentially bound to a column charged with wild-type ClpS and eluted with a peptide bearing an N-degron. Thirty-two of 37 determined N-terminal peptides had N-degrons. Most of the proteins were N-terminally truncated by endoproteases or exopeptidases, and many were further modified by Aat. The identities of the proteins point to possible physiological roles for the N-end rule in cell division, translation, transcription, and DNA replication and reveal widespread proteolytic processing of cellular proteins to generate N-end rule substrates.
Keywords: ATP-dependent Protease; Aat; Adaptor Proteins; ClpA; ClpS; END Site; N-end Rule; Protein Degradation; Protein Processing; Proteomics.
Figures
References
-
- Gottesman S., Wickner S., Maurizi M. R. (1997) Protein quality control: triage by chaperones and proteases. Genes Dev. 11, 815–823 - PubMed
-
- Dougan D. A., Mogk A., Zeth K., Turgay K., Bukau B. (2002) AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529, 6–10 - PubMed
-
- Kessel M., Maurizi M. R., Kim B., Kocsis E., Trus B. L., Singh S. K., Steven A. C. (1995) Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J. Mol. Biol. 250, 587–594 - PubMed
-
- Wang J., Hartling J. A., Flanagan J. M. (1997) The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

