Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase
- PMID: 23960144
- PMCID: PMC3747305
- DOI: 10.1083/jcb.201302123
Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase
Abstract
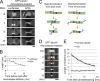
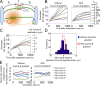
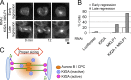
The central spindle is built during anaphase by coupling antiparallel microtubules (MTs) at a central overlap zone, which provides a signaling scaffold for the regulation of cytokinesis. The mechanisms underlying central spindle morphogenesis are still poorly understood. In this paper, we show that the MT depolymerase Kif2A controls the length and alignment of central spindle MTs through depolymerization at their minus ends. The distribution of Kif2A was limited to the distal ends of the central spindle through Aurora B-dependent phosphorylation and exclusion from the spindle midzone. Overactivation or inhibition of Kif2A affected interchromosomal MT length and disorganized the central spindle, resulting in uncoordinated cell division. Experimental data and model simulations suggest that the steady-state length of the central spindle and its symmetric position between segregating chromosomes are predominantly determined by the Aurora B activity gradient. On the basis of these results, we propose a robust self-organization mechanism for central spindle formation.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

