Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824
- PMID: 23962085
- PMCID: PMC3765823
- DOI: 10.1186/1754-6834-6-117
Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824
Abstract
Background: Consolidated bioprocessing (CBP) is reliant on the simultaneous enzyme production, saccharification of biomass, and fermentation of released sugars into valuable products such as butanol. Clostridial species that produce butanol are, however, unable to grow on crystalline cellulose. In contrast, those saccharolytic species that produce predominantly ethanol, such as Clostridium thermocellum and Clostridium cellulolyticum, degrade crystalline cellulose with high efficiency due to their possession of a multienzyme complex termed the cellulosome. This has led to studies directed at endowing butanol-producing species with the genetic potential to produce a cellulosome, albeit by localising the necessary transgenes to unstable autonomous plasmids. Here we have explored the potential of our previously described Allele-Coupled Exchange (ACE) technology for creating strains of the butanol producing species Clostridium acetobutylicum in which the genes encoding the various cellulosome components are stably integrated into the genome.
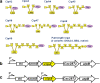
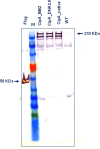
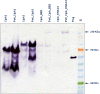
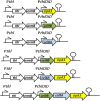
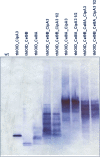
Results: We used BioBrick2 (BB2) standardised parts to assemble a range of synthetic genes encoding C. thermocellum cellulosomal scaffoldin proteins (CipA variants) and glycoside hydrolases (GHs, Cel8A, Cel9B, Cel48S and Cel9K) as well as synthetic cellulosomal operons that direct the synthesis of Cel8A, Cel9B and a truncated form of CipA. All synthetic genes and operons were integrated into the C. acetobutylicum genome using the recently developed ACE technology. Heterologous protein expression levels and mini-cellulosome self-assembly were assayed by western blot and native PAGE analysis.
Conclusions: We demonstrate the successful expression, secretion and self-assembly of cellulosomal subunits by the recombinant C. acetobutylicum strains, providing a platform for the construction of novel cellulosomes.
Figures



Similar articles
-
Stoichiometric Assembly of the Cellulosome Generates Maximum Synergy for the Degradation of Crystalline Cellulose, as Revealed by In Vitro Reconstitution of the Clostridium thermocellum Cellulosome.Appl Environ Microbiol. 2015 Jul;81(14):4756-66. doi: 10.1128/AEM.00772-15. Epub 2015 May 8. Appl Environ Microbiol. 2015. PMID: 25956772 Free PMC article.
-
Revisiting the Regulation of the Primary Scaffoldin Gene in Clostridium thermocellum.Appl Environ Microbiol. 2017 Mar 31;83(8):e03088-16. doi: 10.1128/AEM.03088-16. Print 2017 Apr 15. Appl Environ Microbiol. 2017. PMID: 28159788 Free PMC article.
-
Production of a functional cell wall-anchored minicellulosome by recombinant Clostridium acetobutylicum ATCC 824.Biotechnol Biofuels. 2016 May 23;9:109. doi: 10.1186/s13068-016-0526-x. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 27222664 Free PMC article.
-
Comparative genomics of the mesophilic cellulosome-producing Clostridium cellulovorans and its application to biofuel production via consolidated bioprocessing.Environ Technol. 2010 Jul-Aug;31(8-9):889-903. doi: 10.1080/09593330.2010.490856. Environ Technol. 2010. PMID: 20662379 Review.
-
Biobutanol Production from Crystalline Cellulose through Consolidated Bioprocessing.Trends Biotechnol. 2019 Feb;37(2):167-180. doi: 10.1016/j.tibtech.2018.08.007. Epub 2018 Sep 14. Trends Biotechnol. 2019. PMID: 30224227 Review.
Cited by
-
SBRC-Nottingham: sustainable routes to platform chemicals from C1 waste gases.Biochem Soc Trans. 2016 Jun 15;44(3):684-6. doi: 10.1042/BST20160010. Biochem Soc Trans. 2016. PMID: 27284026 Free PMC article.
-
Improved n-Butanol Production from Clostridium cellulovorans by Integrated Metabolic and Evolutionary Engineering.Appl Environ Microbiol. 2019 Mar 22;85(7):e02560-18. doi: 10.1128/AEM.02560-18. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30658972 Free PMC article.
-
Mutant generation by allelic exchange and genome resequencing of the biobutanol organism Clostridium acetobutylicum ATCC 824.Biotechnol Biofuels. 2016 Jan 4;9:4. doi: 10.1186/s13068-015-0410-0. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 26732067 Free PMC article.
-
A metabolic pathway for bile acid dehydroxylation by the gut microbiome.Nature. 2020 Jun;582(7813):566-570. doi: 10.1038/s41586-020-2396-4. Epub 2020 Jun 17. Nature. 2020. PMID: 32555455 Free PMC article.
-
A roadmap for gene system development in Clostridium.Anaerobe. 2016 Oct;41:104-112. doi: 10.1016/j.anaerobe.2016.05.011. Epub 2016 May 24. Anaerobe. 2016. PMID: 27234263 Free PMC article. Review.
References
-
- Kumar M, Gayen K. Developments in biobutanol production: new insights. Applied Energy. 2011;88:1999–2012. doi: 10.1016/j.apenergy.2010.12.055. - DOI
-
- Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77:8288–8294. doi: 10.1128/AEM.00646-11. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

