Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains
- PMID: 23963180
- PMCID: PMC3747589
- DOI: 10.1128/mBio.00534-13
Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains
Abstract
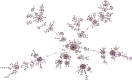
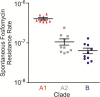
Enterococcus faecium, natively a gut commensal organism, emerged as a leading cause of multidrug-resistant hospital-acquired infection in the 1980s. As the living record of its adaptation to changes in habitat, we sequenced the genomes of 51 strains, isolated from various ecological environments, to understand how E. faecium emerged as a leading hospital pathogen. Because of the scale and diversity of the sampled strains, we were able to resolve the lineage responsible for epidemic, multidrug-resistant human infection from other strains and to measure the evolutionary distances between groups. We found that the epidemic hospital-adapted lineage is rapidly evolving and emerged approximately 75 years ago, concomitant with the introduction of antibiotics, from a population that included the majority of animal strains, and not from human commensal lines. We further found that the lineage that included most strains of animal origin diverged from the main human commensal line approximately 3,000 years ago, a time that corresponds to increasing urbanization of humans, development of hygienic practices, and domestication of animals, which we speculate contributed to their ecological separation. Each bifurcation was accompanied by the acquisition of new metabolic capabilities and colonization traits on mobile elements and the loss of function and genome remodeling associated with mobile element insertion and movement. As a result, diversity within the species, in terms of sequence divergence as well as gene content, spans a range usually associated with speciation.
Importance: Enterococci, in particular vancomycin-resistant Enterococcus faecium, recently emerged as a leading cause of hospital-acquired infection worldwide. In this study, we examined genome sequence data to understand the bacterial adaptations that accompanied this transformation from microbes that existed for eons as members of host microbiota. We observed changes in the genomes that paralleled changes in human behavior. An initial bifurcation within the species appears to have occurred at a time that corresponds to the urbanization of humans and domestication of animals, and a more recent bifurcation parallels the introduction of antibiotics in medicine and agriculture. In response to the opportunity to fill niches associated with changes in human activity, a rapidly evolving lineage emerged, a lineage responsible for the vast majority of multidrug-resistant E. faecium infections.
Figures



References
-
- Arias CA, Murray BE. 2008. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti Infect. Ther. 6:637–655 - PubMed
-
- Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480:241–244 - PubMed
-
- Harrison EM, Paterson GK, Holden MTG, Larsen J, Stegger M, Larsen AR, Petersen A, Skov RL, Christensen JM, Bak Zeuthen A, Heltberg O, Harris SR, Zadoks RN, Parkhill J, Peacock SJ, Holmes MA. 2013. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol. Med. 5:509–515 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
