Transcriptional induction of ADAMTS5 protein by nuclear factor-κB (NF-κB) family member RelA/p65 in chondrocytes during osteoarthritis development
- PMID: 23963448
- PMCID: PMC3789961
- DOI: 10.1074/jbc.M113.452169
Transcriptional induction of ADAMTS5 protein by nuclear factor-κB (NF-κB) family member RelA/p65 in chondrocytes during osteoarthritis development
Abstract
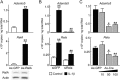
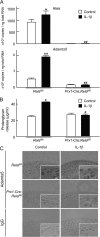
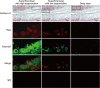
Here we sought to identify transcription factors that induce ADAMTS5, a crucial proteinase for osteoarthritis development. Exhaustive comparison of the genomic sequences of human, macaque, and mouse ADAMTS5 genes revealed that the proximal 1.4 kb of the 5'-end-flanking regions containing several consensus motifs was highly conserved. Among putative transcription factors for these motifs, NF-κB family member RelA/p65 most strongly stimulated the promoter activity. In the ADAMTS5 gene, there were three NF-κB binding motifs, in which deletion, mutagenesis, and tandem repeat analyses of the luciferase assay identified the core responsive elements of RelA/p65 to be -896/-887 and -424/-415 bp with specific bindings. The endogenous Adamts5 expression in ATDC5 cells was increased by RelA/p65 overexpression and decreased by knockdown through its siRNA. The expression was also inhibited by the Rela deletion through Cre transfection in primary articular chondrocytes from Rela(fl/fl) mice. In the ex vivo culture of femoral head cartilage from mesenchymal cell-specific Rela knock-out (Prx1-Cre;Rela(fl/fl)) mice, aggrecanolysis was significantly lower than that in the Rela(fl/fl) cartilage. Finally, in the experimental mouse osteoarthritis model, ADAMTS5 and RelA were co-localized in chondrocytes of degraded articular cartilage. We conclude that RelA/p65 is a potent transcriptional activator of ADAMTS5 in chondrocytes during osteoarthritis development.
Keywords: ADAM ADAMTS; ADAMTS5; Articular Cartilage; Cartilage; Cartilage Biology; Chondrocytes; NF-kappa B (NF-KB); Osteoarthritis; RelA/p65; Transcription.
Figures


References
-
- Verma P., Dalal K. (2011) ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J. Cell. Biochem. 112, 3507–3514 - PubMed
-
- Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., Majumdar M. K., Morris E. A. (2005) Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 434, 644–648 - PubMed
-
- Stanton H., Rogerson F. M., East C. J., Golub S. B., Lawlor K. E., Meeker C. T., Little C. B., Last K., Farmer P. J., Campbell I. K., Fourie A. M., Fosang A. J. (2005) ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 434, 648–652 - PubMed
-
- Glasson S. S., Askew R., Sheppard B., Carito B. A., Blanchet T., Ma H. L., Flannery C. R., Kanki K., Wang E., Peluso D., Yang Z., Majumdar M. K., Morris E. A. (2004) Characterization of and osteoarthritis susceptibility in ADAMTS-4-knockout mice. Arthritis Rheum. 50, 2547–2558 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

