A cargo-centered perspective on the PEX5 receptor-mediated peroxisomal protein import pathway
- PMID: 23963456
- PMCID: PMC3790014
- DOI: 10.1074/jbc.M113.487140
A cargo-centered perspective on the PEX5 receptor-mediated peroxisomal protein import pathway
Abstract
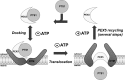
Peroxisomal matrix proteins are synthesized on cytosolic ribosomes and post-translationally targeted to the organelle by PEX5, the peroxisomal shuttling receptor. The pathway followed by PEX5 during this process is known with reasonable detail. After recognizing cargo proteins in the cytosol, the receptor interacts with the peroxisomal docking/translocation machinery, where it gets inserted; PEX5 is then monoubiquitinated, extracted back to the cytosol and, finally, deubiquitinated. However, despite this information, the exact step of this pathway where cargo proteins are translocated across the organelle membrane is still ill-defined. In this work, we used an in vitro import system to characterize the translocation mechanism of a matrix protein possessing a type 1 targeting signal. Our results suggest that translocation of proteins across the organelle membrane occurs downstream of a reversible docking step and upstream of the first cytosolic ATP-dependent step (i.e. before ubiquitination of PEX5), concomitantly with the insertion of the receptor into the docking/translocation machinery.
Keywords: In Vitro Import Assays; PEX5; PTS1 Protein; Peroxisomes; Protein Sorting; Protein Translocation; Subcellular Organelles; Ubiquitination.
Figures




Similar articles
-
Mapping the cargo protein membrane translocation step into the PEX5 cycling pathway.J Biol Chem. 2009 Oct 2;284(40):27243-51. doi: 10.1074/jbc.M109.032565. Epub 2009 Jul 23. J Biol Chem. 2009. PMID: 19632994 Free PMC article.
-
A PEX7-centered perspective on the peroxisomal targeting signal type 2-mediated protein import pathway.Mol Cell Biol. 2014 Aug;34(15):2917-28. doi: 10.1128/MCB.01727-13. Epub 2014 May 27. Mol Cell Biol. 2014. PMID: 24865970 Free PMC article.
-
PEX5 and ubiquitin dynamics on mammalian peroxisome membranes.PLoS Comput Biol. 2014 Jan;10(1):e1003426. doi: 10.1371/journal.pcbi.1003426. Epub 2014 Jan 16. PLoS Comput Biol. 2014. PMID: 24453954 Free PMC article.
-
Protein transport into peroxisomes: Knowns and unknowns.Bioessays. 2017 Oct;39(10). doi: 10.1002/bies.201700047. Epub 2017 Aug 8. Bioessays. 2017. PMID: 28787099 Review.
-
Ubiquitin in the peroxisomal protein import pathway.Biochimie. 2014 Mar;98:29-35. doi: 10.1016/j.biochi.2013.08.003. Epub 2013 Aug 14. Biochimie. 2014. PMID: 23954799 Review.
Cited by
-
Revisiting the intraperoxisomal pathway of mammalian PEX7.Sci Rep. 2015 Jul 3;5:11806. doi: 10.1038/srep11806. Sci Rep. 2015. PMID: 26138649 Free PMC article.
-
The VDAC2-BAK axis regulates peroxisomal membrane permeability.J Cell Biol. 2017 Mar 6;216(3):709-722. doi: 10.1083/jcb.201605002. Epub 2017 Feb 7. J Cell Biol. 2017. PMID: 28174205 Free PMC article.
-
Noncanonical and reversible cysteine ubiquitination prevents the overubiquitination of PEX5 at the peroxisomal membrane.PLoS Biol. 2024 Mar 12;22(3):e3002567. doi: 10.1371/journal.pbio.3002567. eCollection 2024 Mar. PLoS Biol. 2024. PMID: 38470934 Free PMC article.
-
The peroxisomal protein import machinery displays a preference for monomeric substrates.Open Biol. 2015 Apr;5(4):140236. doi: 10.1098/rsob.140236. Open Biol. 2015. PMID: 25854684 Free PMC article.
-
A missense allele of PEX5 is responsible for the defective import of PTS2 cargo proteins into peroxisomes.Hum Genet. 2021 Apr;140(4):649-666. doi: 10.1007/s00439-020-02238-z. Epub 2021 Jan 2. Hum Genet. 2021. PMID: 33389129
References
-
- Lazarow P. B., Fujiki Y. (1985) Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530 - PubMed
-
- Brocard C., Hartig A. (2006) Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 - PubMed
-
- Lazarow P. B. (2006) The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763, 1599–1604 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

