Micro- and nanofluidic technologies for epigenetic profiling
- PMID: 23964309
- PMCID: PMC3739826
- DOI: 10.1063/1.4816835
Micro- and nanofluidic technologies for epigenetic profiling
Abstract
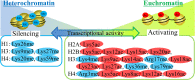
This short review provides an overview of the impact micro- and nanotechnologies can make in studying epigenetic structures. The importance of mapping histone modifications on chromatin prompts us to highlight the complexities and challenges associated with histone mapping, as compared to DNA sequencing. First, the histone code comprised over 30 variations, compared to 4 nucleotides for DNA. Second, whereas DNA can be amplified using polymerase chain reaction, chromatin cannot be amplified, creating challenges in obtaining sufficient material for analysis. Third, while every person has only a single genome, there exist multiple epigenomes in cells of different types and origins. Finally, we summarize existing technologies for performing these types of analyses. Although there are still relatively few examples of micro- and nanofluidic technologies for chromatin analysis, the unique advantages of using such technologies to address inherent challenges in epigenetic studies, such as limited sample material, complex readouts, and the need for high-content screens, make this an area of significant growth and opportunity.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
