Principles of miRNA-target regulation in metazoan models
- PMID: 23965954
- PMCID: PMC3759911
- DOI: 10.3390/ijms140816280
Principles of miRNA-target regulation in metazoan models
Abstract
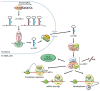
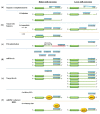
MicroRNAs (miRs) are key post-transcriptional regulators that silence gene expression by direct base pairing to target sites of RNAs. They have a wide variety of tissue expression patterns and are differentially expressed during development and disease. Their activity and abundance is subject to various levels of control ranging from transcription and biogenesis to miR response elements on RNAs, target cellular levels and miR turnover. This review summarizes and discusses current knowledge on the regulation of miR activity and concludes with novel non-canonical functions that have recently emerged.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

