Accidental interaction between PDZ domains and diclofenac revealed by NMR-assisted virtual screening
- PMID: 23966078
- PMCID: PMC6270271
- DOI: 10.3390/molecules18089567
Accidental interaction between PDZ domains and diclofenac revealed by NMR-assisted virtual screening
Abstract
In silico approaches have become indispensable for drug discovery as well as drug repositioning and adverse effect prediction. We have developed the eF-seek program to predict protein-ligand interactions based on the surface structure of proteins using a clique search algorithm. We have also developed a special protein structure prediction pipeline and accumulated predicted 3D models in the Structural Atlas of the Human Genome (SAHG) database. Using this database, genome-wide prediction of non-peptide ligands for proteins in the human genome was performed, and a subset of predicted interactions including 14 PDZ domains was then confirmed by NMR titration. Surprisingly, diclofenac, a non-steroidal anti-inflammatory drug, was found to be a non-peptide PDZ domain ligand, which bound to 5 of 15 tested PDZ domains. The critical residues for the PDZ-diclofenac interaction were also determined. Pharmacological implications of the accidental PDZ-diclofenac interaction are further discussed.
Figures
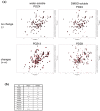
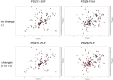

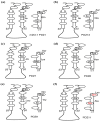

Similar articles
-
Cluster based prediction of PDZ-peptide interactions.BMC Genomics. 2014;15 Suppl 1(Suppl 1):S5. doi: 10.1186/1471-2164-15-S1-S5. Epub 2014 Jan 24. BMC Genomics. 2014. PMID: 24564547 Free PMC article.
-
Structure-based identification of CaMKIIα-interacting MUPP1 PDZ domains and rational design of peptide ligands to target such interaction in human fertilization.Amino Acids. 2016 Jun;48(6):1509-21. doi: 10.1007/s00726-016-2211-6. Epub 2016 Mar 17. Amino Acids. 2016. PMID: 26984442
-
A regression framework incorporating quantitative and negative interaction data improves quantitative prediction of PDZ domain-peptide interaction from primary sequence.Bioinformatics. 2011 Feb 1;27(3):383-90. doi: 10.1093/bioinformatics/btq657. Epub 2010 Dec 2. Bioinformatics. 2011. PMID: 21127034 Free PMC article.
-
Structure function relations in PDZ-domain-containing proteins: Implications for protein networks in cellular signalling.J Biosci. 2018 Mar;43(1):155-171. J Biosci. 2018. PMID: 29485124 Review.
-
Rational Design of PDZ Domain Inhibitors: Discovery of Small Organic Compounds Targeting PDZ Domains.Methods Mol Biol. 2021;2256:277-289. doi: 10.1007/978-1-0716-1166-1_16. Methods Mol Biol. 2021. PMID: 34014528 Review.
Cited by
-
Comparison of SynCAM1/CADM1 PDZ interactions with MUPP1 using mammalian and bacterial pull-down systems.Brain Behav. 2020 Apr;10(4):e01587. doi: 10.1002/brb3.1587. Epub 2020 Feb 28. Brain Behav. 2020. PMID: 32108449 Free PMC article.
-
Storming the gate: New approaches for targeting the dynamic tight junction for improved drug delivery.Adv Drug Deliv Rev. 2023 Aug;199:114905. doi: 10.1016/j.addr.2023.114905. Epub 2023 Jun 3. Adv Drug Deliv Rev. 2023. PMID: 37271282 Free PMC article. Review.
-
NMR-Guided Repositioning of Non-Steroidal Anti-Inflammatory Drugs into Tight Junction Modulators.Int J Mol Sci. 2021 Mar 4;22(5):2583. doi: 10.3390/ijms22052583. Int J Mol Sci. 2021. PMID: 33806674 Free PMC article.
-
Discovery of Potent Disheveled/Dvl Inhibitors Using Virtual Screening Optimized With NMR-Based Docking Performance Index.Front Pharmacol. 2018 Sep 5;9:983. doi: 10.3389/fphar.2018.00983. eCollection 2018. Front Pharmacol. 2018. PMID: 30233369 Free PMC article.
-
Pharmacologic Comparison of High-Dose Hesperetin and Quercetin on MDCK II Cell Viability, Tight Junction Integrity, and Cell Shape.Antioxidants (Basel). 2023 Apr 18;12(4):952. doi: 10.3390/antiox12040952. Antioxidants (Basel). 2023. PMID: 37107328 Free PMC article.
References
-
- Sousa S.F., Fernandes P.A., Ramos M.J. Protein-ligand docking: Current status and future challenges. Proteins. 2006;65:15–26. - PubMed
-
- Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

