Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi
- PMID: 23966099
- PMCID: PMC3906807
- DOI: 10.1038/ismej.2013.143
Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi
Abstract
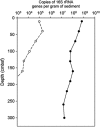
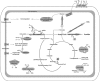
Bacteria of the class Dehalococcoidia (DEH), phylum Chloroflexi, are widely distributed in the marine subsurface, yet metabolic properties of the many uncultivated lineages are completely unknown. This study therefore analysed genomic content from a single DEH cell designated 'DEH-J10' obtained from the sediments of Aarhus Bay, Denmark. Real-time PCR showed the DEH-J10 phylotype was abundant in upper sediments but was absent below 160 cm below sea floor. A 1.44 Mbp assembly was obtained and was estimated to represent up to 60.8% of the full genome. The predicted genome is much larger than genomes of cultivated DEH and appears to confer metabolic versatility. Numerous genes encoding enzymes of core and auxiliary beta-oxidation pathways were identified, suggesting that this organism is capable of oxidising various fatty acids and/or structurally related substrates. Additional substrate versatility was indicated by genes, which may enable the bacterium to oxidise aromatic compounds. Genes encoding enzymes of the reductive acetyl-CoA pathway were identified, which may also enable the fixation of CO2 or oxidation of organics completely to CO2. Genes encoding a putative dimethylsulphoxide reductase were the only evidence for a respiratory terminal reductase. No evidence for reductive dehalogenase genes was found. Genetic evidence also suggests that the organism could synthesise ATP by converting acetyl-CoA to acetate by substrate-level phosphorylation. Other encoded enzymes putatively conferring marine adaptations such as salt tolerance and organo-sulphate sulfohydrolysis were identified. Together, these analyses provide the first insights into the potential metabolic traits that may enable members of the DEH to occupy an ecological niche in marine sediments.
Figures

Similar articles
-
Single-Cell Genome and Group-Specific dsrAB Sequencing Implicate Marine Members of the Class Dehalococcoidia (Phylum Chloroflexi) in Sulfur Cycling.mBio. 2016 May 3;7(3):e00266-16. doi: 10.1128/mBio.00266-16. mBio. 2016. PMID: 27143384 Free PMC article.
-
Homoacetogenesis in Deep-Sea Chloroflexi, as Inferred by Single-Cell Genomics, Provides a Link to Reductive Dehalogenation in Terrestrial Dehalococcoidetes.mBio. 2017 Dec 19;8(6):e02022-17. doi: 10.1128/mBio.02022-17. mBio. 2017. PMID: 29259088 Free PMC article.
-
Development and application of primers for the class Dehalococcoidia (phylum Chloroflexi) enables deep insights into diversity and stratification of subgroups in the marine subsurface.Environ Microbiol. 2015 Oct;17(10):3540-56. doi: 10.1111/1462-2920.12510. Epub 2014 Jun 26. Environ Microbiol. 2015. PMID: 24889097
-
Distribution of Dehalococcoidia in the Anaerobic Deep Water of a Remote Meromictic Crater Lake and Detection of Dehalococcoidia-Derived Reductive Dehalogenase Homologous Genes.PLoS One. 2016 Jan 6;11(1):e0145558. doi: 10.1371/journal.pone.0145558. eCollection 2016. PLoS One. 2016. PMID: 26734727 Free PMC article.
-
Genomic insights into organohalide respiration.Curr Opin Biotechnol. 2013 Jun;24(3):498-505. doi: 10.1016/j.copbio.2013.02.014. Epub 2013 Mar 13. Curr Opin Biotechnol. 2013. PMID: 23490446 Review.
Cited by
-
A comprehensive overview of the Chloroflexota community in wastewater treatment plants worldwide.mSystems. 2023 Dec 21;8(6):e0066723. doi: 10.1128/msystems.00667-23. Epub 2023 Nov 22. mSystems. 2023. PMID: 37992299 Free PMC article. Review.
-
Microbial diversity in a submarine carbonate edifice from the serpentinizing hydrothermal system of the Prony Bay (New Caledonia) over a 6-year period.Front Microbiol. 2015 Aug 27;6:857. doi: 10.3389/fmicb.2015.00857. eCollection 2015. Front Microbiol. 2015. PMID: 26379636 Free PMC article.
-
Vertical Stratification of Dissolved Organic Matter Linked to Distinct Microbial Communities in Subtropic Estuarine Sediments.Front Microbiol. 2021 Jul 20;12:697860. doi: 10.3389/fmicb.2021.697860. eCollection 2021. Front Microbiol. 2021. PMID: 34354693 Free PMC article.
-
Organic Matter Type Defines the Composition of Active Microbial Communities Originating From Anoxic Baltic Sea Sediments.Front Microbiol. 2021 May 5;12:628301. doi: 10.3389/fmicb.2021.628301. eCollection 2021. Front Microbiol. 2021. PMID: 34025597 Free PMC article.
-
Phylogenetically and metabolically diverse autotrophs in the world's deepest blue hole.ISME Commun. 2023 Nov 14;3(1):117. doi: 10.1038/s43705-023-00327-4. ISME Commun. 2023. PMID: 37964026 Free PMC article.
References
-
- Adrian L, Szewzyk U, Wecke J, Görisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. - PubMed
-
- Adrian L. ERC-group microflex: microbiology of Dehalococcoides-like Chloroflexi. Rev Environ Sci Biotechnol. 2009;8:225–229.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

