Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F₁F0-ATP synthase
- PMID: 23966169
- PMCID: PMC3899192
- DOI: 10.4161/cc.25870
Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F₁F0-ATP synthase
Abstract
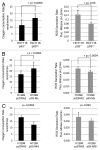
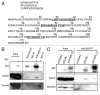
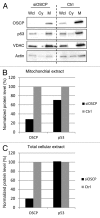
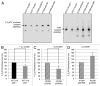
We and others previously reported that endogenous p53 can be located at mitochondria in the absence of stress, suggesting that p53 has a role in the normal physiology of this organelle. The aim of this study was to characterize in unstressed cells the intramitochondrial localization of p53 and identify new partners and functions of p53 in mitochondria. We find that the intramitochondrial pool of p53 is located in the intermembrane space and the matrix. Of note, unstressed HCT116 p53(+/+) cells simultaneously show increased O₂ consumption and decreased mitochondrial superoxide production compared with their p53-null counterpart. This data was confirmed by stable H1299 cell lines expressing low levels of p53 specifically targeted to the matrix. Using immunoprecipitation and mass spectrometry, we identified the oligomycin sensitivity-conferring protein (OSCP), a subunit of the F₁F₀-ATP synthase complex, as a new partner of endogenous p53, specifically interacting with p53 localized in the matrix. Interestingly, this interaction seems implicated in mitochondrial p53 localization. Moreover, p53 localized in the matrix promotes the assembly of F₁F₀-ATP synthase. Taking into account that deregulations of mitochondrial respiration and reactive oxygen species production are tightly linked to cancer development, we suggest that mitochondrial p53 may be an important regulator of normal mitochondrial and cellular physiology, potentially exerting tumor suppression activity inside mitochondria.
Keywords: OSCP; cancer; energetic metabolism; mitochondria; oxidative phosphorylation; p53; reactive oxygen species; respiration; tumor suppressor.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
