A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140
- PMID: 23966389
- PMCID: PMC3807357
- DOI: 10.1128/JVI.01681-13
A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140
Abstract
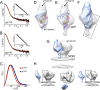
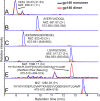
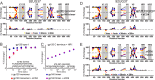
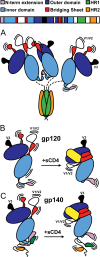
The envelope glycoprotein (Env) is the sole antigenic feature on the surface of HIV and the target for the humoral immune system. Soluble, uncleaved gp140 Env constructs truncated at the transmembrane domain are being investigated intensively as potential vaccine immunogens by many groups, and understanding their structural properties is essential. We used hydrogen/deuterium-exchange mass spectrometry and small-angle X-ray scattering to probe structural order in a panel of commonly used gp140 constructs and matched gp120 monomers. We observed that oligomeric forms of uncleaved gp140, generally presumed to be trimeric, contain a protease-resistant form of gp41 akin to the postfusion, helical bundle conformation and appear to lack specific interactions between gp120 and gp41. In contrast, the monomeric form of gp140 shows significant stabilization of the gp120 inner domain imparted by the gp41 region, demonstrating excellent agreement with past mutagenesis studies. Moreover, the gp140 monomers respond to CD4 binding in manner that is consistent with the initial stages of Env activation: CD4 binding induces structural ordering throughout gp120 while loosening its association with gp41. The results indicate that uncleaved gp140 oligomers do not represent an authentic prefusion form of Env, whereas gp140 monomers isolated from the same glycoprotein preparations in many ways exhibit function and internal structural order that are consistent with expectations for certain aspects of native Env. gp140 monomers may thus be a useful reagent for advancing structural and functional studies.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

