Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG
- PMID: 23966868
- PMCID: PMC3744422
- DOI: 10.1371/journal.pgen.1003683
Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG
Abstract
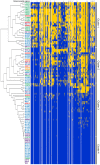
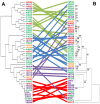
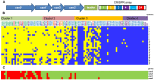
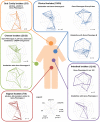
Lactobacillus rhamnosus is a lactic acid bacterium that is found in a large variety of ecological habitats, including artisanal and industrial dairy products, the oral cavity, intestinal tract or vagina. To gain insights into the genetic complexity and ecological versatility of the species L. rhamnosus, we examined the genomes and phenotypes of 100 L. rhamnosus strains isolated from diverse sources. The genomes of 100 L. rhamnosus strains were mapped onto the L. rhamnosus GG reference genome. These strains were phenotypically characterized for a wide range of metabolic, antagonistic, signalling and functional properties. Phylogenomic analysis showed multiple groupings of the species that could partly be associated with their ecological niches. We identified 17 highly variable regions that encode functions related to lifestyle, i.e. carbohydrate transport and metabolism, production of mucus-binding pili, bile salt resistance, prophages and CRISPR adaptive immunity. Integration of the phenotypic and genomic data revealed that some L. rhamnosus strains possibly resided in multiple niches, illustrating the dynamics of bacterial habitats. The present study showed two distinctive geno-phenotypes in the L. rhamnosus species. The geno-phenotype A suggests an adaptation to stable nutrient-rich niches, i.e. milk-derivative products, reflected by the alteration or loss of biological functions associated with antimicrobial activity spectrum, stress resistance, adaptability and fitness to a distinctive range of habitats. In contrast, the geno-phenotype B displays adequate traits to a variable environment, such as the intestinal tract, in terms of nutrient resources, bacterial population density and host effects.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Rajilić-Stojanović M, Smidt H, De Vos WM (2007) Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 9: 2125–2136. - PubMed
-
- Williamson SJ, Yooseph S (2011) From bacterial to microbial ecosystems (metagenomics) Bacterial Molecular Networks. Meth Mol Biol 35–55. - PubMed
-
- Vaughan EE, Schut F, Heilig HG, Zoetendal EG, de Vos WM, et al. (2000) A molecular view of the intestinal ecosystem. Curr Issues Intest Microbiol 1: 1–12. - PubMed
-
- Kleerebezem M, Vaughan EE (2009) Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu Rev Microbiol 63: 269–290. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

