Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment
- PMID: 23967097
- PMCID: PMC3742674
- DOI: 10.1371/journal.pone.0070749
Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment
Abstract
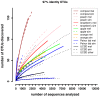
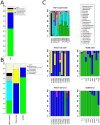
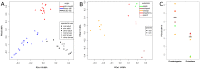
The fruit fly Drosophila is a classic model organism to study adaptation as well as the relationship between genetic variation and phenotypes. Although associated bacterial communities might be important for many aspects of Drosophila biology, knowledge about their diversity, composition, and factors shaping them is limited. We used 454-based sequencing of a variable region of the bacterial 16S ribosomal RNA gene to characterize the bacterial communities associated with wild and laboratory Drosophila isolates. In order to specifically investigate effects of food source and host species on bacterial communities, we analyzed samples from wild Drosophila melanogaster and D. simulans collected from a variety of natural substrates, as well as from adults and larvae of nine laboratory-reared Drosophila species. We find no evidence for host species effects in lab-reared flies; instead, lab of origin and stochastic effects, which could influence studies of Drosophila phenotypes, are pronounced. In contrast, the natural Drosophila-associated microbiota appears to be predominantly shaped by food substrate with an additional but smaller effect of host species identity. We identify a core member of this natural microbiota that belongs to the genus Gluconobacter and is common to all wild-caught flies in this study, but absent from the laboratory. This makes it a strong candidate for being part of what could be a natural D. melanogaster and D. simulans core microbiome. Furthermore, we were able to identify candidate pathogens in natural fly isolates.
Conflict of interest statement
Figures

Similar articles
-
Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system.PLoS Genet. 2011 Sep;7(9):e1002272. doi: 10.1371/journal.pgen.1002272. Epub 2011 Sep 22. PLoS Genet. 2011. PMID: 21966276 Free PMC article.
-
A Distinctive and Host-Restricted Gut Microbiota in Populations of a Cactophilic Drosophila Species.Appl Environ Microbiol. 2017 Nov 16;83(23):e01551-17. doi: 10.1128/AEM.01551-17. Print 2017 Dec 1. Appl Environ Microbiol. 2017. PMID: 28939605 Free PMC article.
-
The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis.ISME J. 2013 Oct;7(10):1922-32. doi: 10.1038/ismej.2013.86. Epub 2013 May 30. ISME J. 2013. PMID: 23719154 Free PMC article.
-
Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster.mBio. 2014 Sep 30;5(5):e01631-14. doi: 10.1128/mBio.01631-14. mBio. 2014. PMID: 25271286 Free PMC article.
-
Stable association of a Drosophila-derived microbiota with its animal partner and the nutritional environment throughout a fly population's life cycle.J Insect Physiol. 2018 Apr;106(Pt 1):2-12. doi: 10.1016/j.jinsphys.2017.09.003. Epub 2017 Sep 12. J Insect Physiol. 2018. PMID: 28916161 Review.
Cited by
-
Probiotic Bifidobacteria Mitigate the Deleterious Effects of para-Cresol in a Drosophila melanogaster Toxicity Model.mSphere. 2022 Dec 21;7(6):e0044622. doi: 10.1128/msphere.00446-22. Epub 2022 Nov 2. mSphere. 2022. PMID: 36321825 Free PMC article.
-
Microbiota disruption leads to reduced cold tolerance in Drosophila flies.Naturwissenschaften. 2018 Sep 17;105(9-10):59. doi: 10.1007/s00114-018-1584-7. Naturwissenschaften. 2018. PMID: 30291448
-
Bacterial Metabolism and Transport Genes Are Associated with the Preference of Drosophila melanogaster for Dietary Yeast.Appl Environ Microbiol. 2022 Aug 23;88(16):e0072022. doi: 10.1128/aem.00720-22. Epub 2022 Aug 1. Appl Environ Microbiol. 2022. PMID: 35913151 Free PMC article.
-
Beyond canonical models: why a broader understanding of Diptera-microbiota interactions is essential for vector-borne disease control.Evol Ecol. 2023 Feb;37(1):165-188. doi: 10.1007/s10682-022-10197-2. Epub 2022 Jul 28. Evol Ecol. 2023. PMID: 37153630 Free PMC article.
-
Microbiome Structure of a Wild Drosophila Community along Tropical Elevational Gradients and Comparison to Laboratory Lines.Appl Environ Microbiol. 2023 May 31;89(5):e0009923. doi: 10.1128/aem.00099-23. Epub 2023 May 8. Appl Environ Microbiol. 2023. PMID: 37154737 Free PMC article.
References
-
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, et al. (2012) Host-Gut Microbiota Metabolic Interactions. Science 336: 1262–1267 doi:10.1126/science.1223813 - DOI - PubMed
-
- Krause DO, Denman SE, Mackie RI, Morrison M, Rae AL, et al. (2003) Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiology Reviews 27: 663–693 doi:10.1016/S0168-6445(03)00072-X - DOI - PubMed
-
- McFall-Ngai M, Heath-Heckman EAC, Gillette AA, Peyer SM, Harvie EA (2012) The secret languages of coevolved symbioses: Insights from the Euprymna scolopes–Vibrio fischeri symbiosis. Seminars in Immunology 24: 3–8 doi:10.1016/j.smim.2011.11.006 - DOI - PMC - PubMed
-
- Kaltenpoth M, Göttler W, Herzner G, Strohm E (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Current Biology 15: 475–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

