The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults
- PMID: 23967406
- PMCID: PMC3744036
- DOI: 10.3389/fcimb.2013.00041
The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults
Abstract
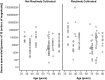
The urinary microbiome of healthy individuals and the way it alters with ageing have not been characterized and may influence disease processes. Conventional microbiological methods have limited scope to capture the full spectrum of urinary bacterial species. We studied the urinary microbiota from a population of healthy individuals, ranging from 26 to 90 years of age, by amplification of the 16S rRNA gene, with resulting amplicons analyzed by 454 pyrosequencing. Mid-stream urine (MSU) was collected by the "clean-catch" method. Quantitative PCR of 16S rRNA genes in urine samples, allowed relative enumeration of the bacterial loads. Analysis of the samples indicates that females had a more heterogeneous mix of bacterial genera compared to the male samples and generally had representative members of the phyla Actinobacteria and Bacteroidetes. Analysis of the data leads us to conclude that a "core" urinary microbiome could potentially exist, when samples are grouped by age with fluctuation in abundance between age groups. The study also revealed age-specific genera Jonquetella, Parvimonas, Proteiniphilum, and Saccharofermentans. In conclusion, conventional microbiological methods are inadequate to fully identify around two-thirds of the bacteria identified in this study. Whilst this proof-of-principle study has limitations due to the sample size, the discoveries evident in this sample data are strongly suggestive that a larger study on the urinary microbiome should be encouraged and that the identification of specific genera at particular ages may be relevant to pathogenesis of clinical conditions.
Keywords: bladder disease; bladder microbiome; microbiological methods; pyrosequencing; urinary microbiome.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

