DNorm: disease name normalization with pairwise learning to rank
- PMID: 23969135
- PMCID: PMC3810844
- DOI: 10.1093/bioinformatics/btt474
DNorm: disease name normalization with pairwise learning to rank
Abstract
Motivation: Despite the central role of diseases in biomedical research, there have been much fewer attempts to automatically determine which diseases are mentioned in a text-the task of disease name normalization (DNorm)-compared with other normalization tasks in biomedical text mining research.
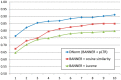
Methods: In this article we introduce the first machine learning approach for DNorm, using the NCBI disease corpus and the MEDIC vocabulary, which combines MeSH® and OMIM. Our method is a high-performing and mathematically principled framework for learning similarities between mentions and concept names directly from training data. The technique is based on pairwise learning to rank, which has not previously been applied to the normalization task but has proven successful in large optimization problems for information retrieval.
Results: We compare our method with several techniques based on lexical normalization and matching, MetaMap and Lucene. Our algorithm achieves 0.782 micro-averaged F-measure and 0.809 macro-averaged F-measure, an increase over the highest performing baseline method of 0.121 and 0.098, respectively.
Availability: The source code for DNorm is available at http://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/DNorm, along with a web-based demonstration and links to the NCBI disease corpus. Results on PubMed abstracts are available in PubTator: http://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/PubTator .
Figures


References
-
- Bai B, et al. Learning to rank with (a lot of) word features. Inf. Retr. 2010;13:291–314.
-
- Biesecker LG. Mapping phenotypes to language: a proposal to organize and standardize the clinical descriptions of malformations. Clin. Genet. 2005;68:320–326. - PubMed
-
- Burges C, et al. Proceedings of the 22nd International Conference on Machine learning. New York, NY, USA: ACM; 2005. Learning to rank using gradient descent; pp. 89–96.
-
- Buyko E, et al. Proceedings of the 10th Conference of the Pacific Association for Computational Linguistics. Melbourne: Pacific Association for Computational Linguistics; 2007. Resolution of coordination ellipses in biological named entities using conditional random fields; pp. 163–171.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

